The CALIS Procedure
-
Overview

-
Getting Started

-
Syntax
 Classes of Statements in PROC CALISSingle-Group Analysis SyntaxMultiple-Group Multiple-Model Analysis SyntaxPROC CALIS StatementBOUNDS StatementBY StatementCOSAN StatementCOV StatementDETERM StatementEFFPART StatementFACTOR StatementFITINDEX StatementFREQ StatementGROUP StatementLINCON StatementLINEQS StatementLISMOD StatementLMTESTS StatementMATRIX StatementMEAN StatementMODEL StatementMSTRUCT StatementNLINCON StatementNLOPTIONS StatementOUTFILES StatementPARAMETERS StatementPARTIAL StatementPATH StatementPATHDIAGRAM StatementPCOV StatementPVAR StatementRAM StatementREFMODEL StatementRENAMEPARM StatementSAS Programming StatementsSIMTESTS StatementSTD StatementSTRUCTEQ StatementTESTFUNC StatementVAR StatementVARIANCE StatementVARNAMES StatementWEIGHT Statement
Classes of Statements in PROC CALISSingle-Group Analysis SyntaxMultiple-Group Multiple-Model Analysis SyntaxPROC CALIS StatementBOUNDS StatementBY StatementCOSAN StatementCOV StatementDETERM StatementEFFPART StatementFACTOR StatementFITINDEX StatementFREQ StatementGROUP StatementLINCON StatementLINEQS StatementLISMOD StatementLMTESTS StatementMATRIX StatementMEAN StatementMODEL StatementMSTRUCT StatementNLINCON StatementNLOPTIONS StatementOUTFILES StatementPARAMETERS StatementPARTIAL StatementPATH StatementPATHDIAGRAM StatementPCOV StatementPVAR StatementRAM StatementREFMODEL StatementRENAMEPARM StatementSAS Programming StatementsSIMTESTS StatementSTD StatementSTRUCTEQ StatementTESTFUNC StatementVAR StatementVARIANCE StatementVARNAMES StatementWEIGHT Statement -
Details
 Input Data SetsOutput Data SetsDefault Analysis Type and Default ParameterizationThe COSAN ModelThe FACTOR ModelThe LINEQS ModelThe LISMOD Model and SubmodelsThe MSTRUCT ModelThe PATH ModelThe RAM ModelNaming Variables and ParametersSetting Constraints on ParametersAutomatic Variable SelectionPath Diagrams: Layout Algorithms, Default Settings, and CustomizationEstimation CriteriaRelationships among Estimation CriteriaGradient, Hessian, Information Matrix, and Approximate Standard ErrorsCounting the Degrees of FreedomAssessment of FitCase-Level Residuals, Outliers, Leverage Observations, and Residual DiagnosticsLatent Variable ScoresTotal, Direct, and Indirect EffectsStandardized SolutionsModification IndicesMissing Values and the Analysis of Missing PatternsMeasures of Multivariate KurtosisInitial EstimatesUse of Optimization TechniquesComputational ProblemsDisplayed OutputODS Table NamesODS Graphics
Input Data SetsOutput Data SetsDefault Analysis Type and Default ParameterizationThe COSAN ModelThe FACTOR ModelThe LINEQS ModelThe LISMOD Model and SubmodelsThe MSTRUCT ModelThe PATH ModelThe RAM ModelNaming Variables and ParametersSetting Constraints on ParametersAutomatic Variable SelectionPath Diagrams: Layout Algorithms, Default Settings, and CustomizationEstimation CriteriaRelationships among Estimation CriteriaGradient, Hessian, Information Matrix, and Approximate Standard ErrorsCounting the Degrees of FreedomAssessment of FitCase-Level Residuals, Outliers, Leverage Observations, and Residual DiagnosticsLatent Variable ScoresTotal, Direct, and Indirect EffectsStandardized SolutionsModification IndicesMissing Values and the Analysis of Missing PatternsMeasures of Multivariate KurtosisInitial EstimatesUse of Optimization TechniquesComputational ProblemsDisplayed OutputODS Table NamesODS Graphics -
Examples
 Estimating Covariances and CorrelationsEstimating Covariances and Means SimultaneouslyTesting Uncorrelatedness of VariablesTesting Covariance PatternsTesting Some Standard Covariance Pattern HypothesesLinear Regression ModelMultivariate Regression ModelsMeasurement Error ModelsTesting Specific Measurement Error ModelsMeasurement Error Models with Multiple PredictorsMeasurement Error Models Specified As Linear EquationsConfirmatory Factor ModelsConfirmatory Factor Models: Some VariationsResidual Diagnostics and Robust EstimationThe Full Information Maximum Likelihood MethodComparing the ML and FIML EstimationPath Analysis: Stability of AlienationSimultaneous Equations with Mean Structures and Reciprocal PathsFitting Direct Covariance StructuresConfirmatory Factor Analysis: Cognitive AbilitiesTesting Equality of Two Covariance Matrices Using a Multiple-Group AnalysisTesting Equality of Covariance and Mean Matrices between Independent GroupsIllustrating Various General Modeling LanguagesTesting Competing Path Models for the Career Aspiration DataFitting a Latent Growth Curve ModelHigher-Order and Hierarchical Factor ModelsLinear Relations among Factor LoadingsMultiple-Group Model for Purchasing BehaviorFitting the RAM and EQS Models by the COSAN Modeling LanguageSecond-Order Confirmatory Factor AnalysisLinear Relations among Factor Loadings: COSAN Model SpecificationOrdinal Relations among Factor LoadingsLongitudinal Factor Analysis
Estimating Covariances and CorrelationsEstimating Covariances and Means SimultaneouslyTesting Uncorrelatedness of VariablesTesting Covariance PatternsTesting Some Standard Covariance Pattern HypothesesLinear Regression ModelMultivariate Regression ModelsMeasurement Error ModelsTesting Specific Measurement Error ModelsMeasurement Error Models with Multiple PredictorsMeasurement Error Models Specified As Linear EquationsConfirmatory Factor ModelsConfirmatory Factor Models: Some VariationsResidual Diagnostics and Robust EstimationThe Full Information Maximum Likelihood MethodComparing the ML and FIML EstimationPath Analysis: Stability of AlienationSimultaneous Equations with Mean Structures and Reciprocal PathsFitting Direct Covariance StructuresConfirmatory Factor Analysis: Cognitive AbilitiesTesting Equality of Two Covariance Matrices Using a Multiple-Group AnalysisTesting Equality of Covariance and Mean Matrices between Independent GroupsIllustrating Various General Modeling LanguagesTesting Competing Path Models for the Career Aspiration DataFitting a Latent Growth Curve ModelHigher-Order and Hierarchical Factor ModelsLinear Relations among Factor LoadingsMultiple-Group Model for Purchasing BehaviorFitting the RAM and EQS Models by the COSAN Modeling LanguageSecond-Order Confirmatory Factor AnalysisLinear Relations among Factor Loadings: COSAN Model SpecificationOrdinal Relations among Factor LoadingsLongitudinal Factor Analysis - References
Gradient, Hessian, Information Matrix, and Approximate Standard Errors
For a single-sample setting with a discrepancy function 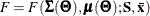 , the gradient is defined as the first partial derivatives of the discrepancy function with respect to the model parameters
, the gradient is defined as the first partial derivatives of the discrepancy function with respect to the model parameters
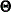 :
:
![\[ g(\bTheta ) = \frac{\partial }{\partial \bTheta } F(\bTheta ) \]](images/statug_calis0592.png)
The Hessian is defined as the second partial derivatives of the discrepancy function with respect to the model parameters
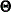 :
:
![\[ H(\bTheta ) = \frac{\partial ^2 }{\partial \bTheta \partial \bTheta ^{\prime }} F(\bTheta ) \]](images/statug_calis0593.png)
Suppose that the mean and covariance structures fit perfectly with 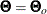 in the population. The expected information matrix is defined as
in the population. The expected information matrix is defined as
![\[ I(\bTheta _ o) = \frac{1}{2} \mathcal{E}(H(\bTheta _ o)) \]](images/statug_calis0595.png)
where the expectation 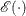 is taken over the sampling space of
is taken over the sampling space of 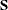 and
and  . Hence, the expected information matrix
. Hence, the expected information matrix 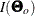 does not contain any sample values.
does not contain any sample values.
The expected information matrix plays a significant role in statistical theory. Under certain regularity conditions, the inverse
of the information matrix 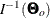 is the asymptotic covariance matrix for
is the asymptotic covariance matrix for 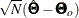 , where N denotes the sample size and
, where N denotes the sample size and 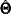 is an estimator.
is an estimator.
In practice, 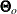 is never known and can only be estimated. The information matrix is therefore evaluating at the sample estimate
is never known and can only be estimated. The information matrix is therefore evaluating at the sample estimate 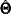 and is denoted as
and is denoted as
![\[ I(\hat{\bTheta }) \]](images/statug_calis0602.png)
This is the information matrix that PROC CALIS displays in the output.
For a sample of size N, PROC CALIS computes the estimated covariance matrix of 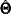 by
by
![\[ ((N-1)I(\hat{\bTheta }))^{-1} \]](images/statug_calis0603.png)
It then computes approximate standard errors for 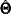 as the square roots of the diagonal elements of this estimated covariance matrix. This formula is based on the expected information
and is the default standard error method (INFORMATION=EXP
) for the ML, MLSB, GLS, and WLS estimation methods.
as the square roots of the diagonal elements of this estimated covariance matrix. This formula is based on the expected information
and is the default standard error method (INFORMATION=EXP
) for the ML, MLSB, GLS, and WLS estimation methods.
In contrast, by default the FIML estimation method computes standard error estimates based on the so-called observed information matrix (INFORMATION=OBS ), which is defined as
![\[ I_{\mathit{obs}}(\bTheta _ o) = \frac{1}{2} H(\bTheta _ o) \]](images/statug_calis0604.png)
The critical difference between 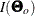 and
and 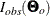 is that the latter does not take the expectation of
is that the latter does not take the expectation of 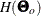 over the distribution of sample statistics. Kenward and Molenberghs (1998) show that the use of the expected information leads to biased standard errors when the missing data mechanism satisfies
only the missing at random (MAR; see Rubin 1976) condition but not the missing completely at random (MCAR) condition. Under the MAR condition, the observed information matrix
is the correct choice. Because the FIML estimation is mostly applied when the data contain missing values, using the observed
information by default is quite reasonable.
over the distribution of sample statistics. Kenward and Molenberghs (1998) show that the use of the expected information leads to biased standard errors when the missing data mechanism satisfies
only the missing at random (MAR; see Rubin 1976) condition but not the missing completely at random (MCAR) condition. Under the MAR condition, the observed information matrix
is the correct choice. Because the FIML estimation is mostly applied when the data contain missing values, using the observed
information by default is quite reasonable.
In practice, the observed information is computed by
![\[ I_{\mathit{obs}}(\hat{\bTheta }) \]](images/statug_calis0607.png)
and the estimated covariance matrix of 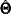 is given by
is given by
![\[ ((N-1)I_{\mathit{obs}}(\hat{\bTheta }))^{-1} \]](images/statug_calis0608.png)
However, PROC CALIS does not compute 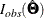 analytically. It computes
analytically. It computes 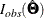 by the finite-difference method based on the analytic formulas of the first-order partial derivatives of F.
by the finite-difference method based on the analytic formulas of the first-order partial derivatives of F.
Finally, PROC CALIS does not compute standard errors when you use the ULS and DWLS estimation methods.
If a particular information matrix is singular, PROC CALIS offers two ways to compute a generalized inverse of the matrix and, therefore, two ways to compute approximate standard errors of implicitly constrained parameter estimates, t values , and modification indices . Depending on the G4= specification, either a Moore-Penrose inverse or a G2 inverse is computed. The computationally expensive Moore-Penrose inverse calculates an estimate of the null space by using an eigenvalue decomposition. The computationally cheaper G2 inverse is produced by sweeping the linearly independent rows and columns and zeroing out the dependent ones.
Satorra-Bentler Sandwich Formula for Standard Errors
In addition to the scaled chi-square statistics, Satorra and Bentler (1994) propose the so-called sandwich formula for computing standard errors. For ML estimation, let 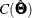 be the estimated covariance matrix of the parameter estimates, obtained through either the expected or observed information
matrix formula. The Satorra-Bentler sandwich formula for the estimated covariance matrix is of the form
be the estimated covariance matrix of the parameter estimates, obtained through either the expected or observed information
matrix formula. The Satorra-Bentler sandwich formula for the estimated covariance matrix is of the form
![\[ C_{\mathit{SB}}(\hat{\bTheta }) = C(\hat{\bTheta })\bUpsilon (\hat{\bSigma })C(\hat{\bTheta }) \]](images/statug_calis0611.png)
where 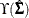 depends on the model Jacobian, the weight matrix under the normal distribution theory, and the weight matrix under general
distribution theory—all evaluated at the sample estimates or the sample data values. See Satorra and Bentler (1994) for detailed formulas.
depends on the model Jacobian, the weight matrix under the normal distribution theory, and the weight matrix under general
distribution theory—all evaluated at the sample estimates or the sample data values. See Satorra and Bentler (1994) for detailed formulas.
If you specify METHOD= MLSB, PROC CALIS uses the Satorra-Bentler sandwich formula to compute standard error estimates. For all other estimation methods that can produce standard error estimates, it uses the unadjusted formula by default. To use the unadjusted formula for METHOD=MLSB, you can specify the SE= UNADJ option. To use the Satorra-Bentler sandwich formula for regular ML estimation, you can specify the SE= SBSW option.
Theoretically, if the population is truly multivariate normal, the weight matrix under normal distribution theory is correctly
specified. Asymptotically, the first two terms in the formula for 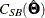 cancel out, so that
cancel out, so that
![\[ C_{\mathit{SB}}(\hat{\bTheta }) = C(\hat{\bTheta }) \]](images/statug_calis0614.png)
That is, you can use the unadjusted covariance formula to compute standard error estimates if the multivariate normality assumption is satisfied.
If the multivariate normal assumption is not true, then the full sandwich formula has to be involved. Specifically, the middle
term, 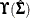 , in the sandwich formula needs to compute the normal-theory weight matrix and other quantities. Because the normal-theory
weight matrix is a function of
, in the sandwich formula needs to compute the normal-theory weight matrix and other quantities. Because the normal-theory
weight matrix is a function of 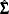 , evaluation of
, evaluation of 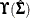 depends on the choice of
depends on the choice of 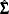 . PROC CALIS uses the model-predicted covariance matrix by default (SBNTW=
PRED). You can also use the sample covariance matrix by specifying the SBNTW=
OBS option.
. PROC CALIS uses the model-predicted covariance matrix by default (SBNTW=
PRED). You can also use the sample covariance matrix by specifying the SBNTW=
OBS option.
Multiple-Group Extensions
In the section Multiple-Group Discrepancy Function, the overall discrepancy function for multiple-group analysis is defined. The same notation is applied here. To begin with,
the overall discrepancy function 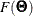 is expressed as a weighted sum of individual discrepancy functions
is expressed as a weighted sum of individual discrepancy functions 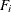 ’s for the groups as follows:
’s for the groups as follows:
![\[ F(\bTheta ) = \sum _{i=1}^ k t_ i F_ i(\bTheta ) \]](images/statug_calis0532.png)
where
![\[ t_ i = \frac{N_ i-1}{N-k} \]](images/statug_calis0533.png)
is the weight for group i,
![\[ N = \sum _{i=1}^ k N_ i \]](images/statug_calis0534.png)
is the total sample size, and 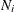 is the sample size for group i.
is the sample size for group i.
The gradient 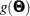 and the Hessian
and the Hessian 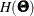 are now defined as weighted sum of individual functions. That is,
are now defined as weighted sum of individual functions. That is,
![\[ g(\bTheta ) = \sum _{i=1}^ k t_ i g_ i(\bTheta ) = \sum _{i=1}^ k t_ i \frac{\partial }{\partial \bTheta } F_ i(\bTheta ) \]](images/statug_calis0619.png)
and
![\[ H(\bTheta ) = \sum _{i=1}^ k t_ i H_ i(\bTheta ) = \sum _{i=1}^ k t_ i \frac{\partial ^2 }{\partial \bTheta \partial \bTheta ^{\prime }} F_ i(\bTheta ) \]](images/statug_calis0620.png)
Suppose that the mean and covariance structures fit perfectly with 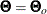 in the population. If each
in the population. If each 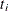 converges to a fixed constant
converges to a fixed constant 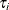 (
(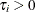 ) with increasing total sample size, the expected information matrix can be written as:
) with increasing total sample size, the expected information matrix can be written as:
![\[ I(\bTheta _ o) = \frac{1}{2} \sum _{i=1}^ k \tau _ i \mathcal{E}(H_ i(\bTheta _ o)) \]](images/statug_calis0624.png)
To compute the expected information empirically, 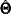 replaces
replaces 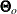 in the formula.
in the formula.
PROC CALIS computes the estimated covariance matrix of 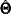 by:
by:
![\[ ((N-k)I(\hat{\bTheta }))^{-1} \]](images/statug_calis0625.png)
Approximate standard errors for 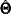 are then computed as the square roots of the diagonal elements of this estimated covariance matrix.
are then computed as the square roots of the diagonal elements of this estimated covariance matrix.
Again, by default the ML, MLSB, GLS, and WLS estimation use such an expected-information based method in the multiple-group setting. For the FIML estimation, the default standard error method is based on the observed information, which is defined as:
![\[ I_{\mathit{obs}}(\bTheta _ o) = \frac{1}{2} \sum _{i=1}^ k \tau _ i H_ i(\bTheta _ o) \]](images/statug_calis0626.png)
Similar to the single-group analysis, standard errors are not computed with the ULS and DWLS estimation methods in the multiple-group setting.
Testing Rank Deficiency in the Approximate Covariance Matrix for Parameter Estimates
When computing the approximate covariance matrix and hence the standard errors for the parameter estimates, inversion of the scaled information matrix or Hessian matrix is involved. The numerical condition of the information matrix can be very poor in many practical applications, especially for the analysis of unscaled covariance data. The following four-step strategy is used for the inversion of the information matrix.
-
The inversion (usually of a normalized matrix
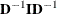 ) is tried using a modified form of the Bunch and Kaufman (1977) algorithm, which allows the specification of a different singularity criterion for each pivot. The following three criteria
for the detection of rank loss in the information matrix are used to specify thresholds:
) is tried using a modified form of the Bunch and Kaufman (1977) algorithm, which allows the specification of a different singularity criterion for each pivot. The following three criteria
for the detection of rank loss in the information matrix are used to specify thresholds:
-
ASING specifies absolute singularity.
-
MSING specifies relative singularity depending on the whole matrix norm.
-
VSING specifies relative singularity depending on the column matrix norm.
If no rank loss is detected, the inverse of the information matrix is used for the covariance matrix of parameter estimates, and the next two steps are skipped.
-
-
The linear dependencies among the parameter subsets are displayed based on the singularity criteria.
-
If the number of parameters t is smaller than the value specified by the G4= option (the default value is 60), the Moore-Penrose inverse is computed based on the eigenvalue decomposition of the information matrix. If you do not specify the NOPRINT option, the distribution of eigenvalues is displayed, and those eigenvalues that are set to zero in the Moore-Penrose inverse are indicated. You should inspect this eigenvalue distribution carefully.
-
If PROC CALIS did not set the right subset of eigenvalues to zero, you can specify the COVSING= option to set a larger or smaller subset of eigenvalues to zero in a further run of PROC CALIS.