The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
How PROC MCMC Works
PROC MCMC is a simulation-based procedure that applies a variety of sampling algorithms to the program at hand. The default sampling methods include conjugate sampling (from full conditional), direct sampling from the marginal distribution, inverse cumulative distribution function, random walk Metropolis with normal proposal, and discretized random walk Metropolis with normal proposal. You can request alternate sampling algorithms, such as random walk Metropolis with t distribution proposal, discretized random walk Metropolis with symmetric geometric proposal, and the slice sampling algorithm.
PROC MCMC applies the more efficient sampling algorithms first, whenever possible. When a parameter does not appear in the conditional distributions of other random variables in the program, PROC MCMC generates samples directly from its prior distribution (which is also its marginal distribution). This usually occurs in data-independent Monte Carlo simulation programs (see Simulating Samples From a Known Density for an example) or missing data problems, where the missing response variables are generated directly from the conditional sampling distribution (or the conditional likelihood). When conjugacy is detected, PROC MCMC uses random number generators to draw values from the full conditional distribution. (For information about detecting conjugacy, see the section Conjugate Sampling.) In other situations, PROC MCMC resorts to the random walk Metropolis with normal proposal to generate posterior samples for continuous parameters and a discretized version for discrete parameters. See the section Metropolis and Metropolis-Hastings Algorithms in Chapter 7: Introduction to Bayesian Analysis Procedures, for details about the Metropolis algorithm. For the actual implementation details of the Metropolis algorithm in PROC MCMC, such as tuning of the covariance matrices, see the section Tuning the Proposal Distribution.
A key component of the Metropolis algorithm is the calculation of the objective function. In most cases, the objective function that PROC MCMC uses in a Metropolis step is the logarithm of the joint posterior distribution, which is calculated with the inclusion of all data and parameters. The rest of this section describes how PROC MCMC calculates the objective function for parameters that use the Metropolis algorithm.
Model Parameters
To calculate the log of the posterior density, PROC MCMC assumes that all observations in the data set are independent,
![\[ \log (p(\theta | \mb{y})) = \log (\pi (\theta )) + \sum _{i=1}^ n \log (f(y_ i|\theta )) \]](images/statug_mcmc0114.png)
where 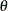 is a parameter or a vector of parameters that are defined in the PARMS
statements (referred to as the model parameters). The term
is a parameter or a vector of parameters that are defined in the PARMS
statements (referred to as the model parameters). The term 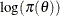 is the sum of the log of the prior densities specified in the PRIOR
and HYPERPRIOR
statements. The term
is the sum of the log of the prior densities specified in the PRIOR
and HYPERPRIOR
statements. The term 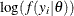 is the log likelihood specified in the MODEL
statement. The MODEL
statement specifies the log likelihood for a single observation in the data set.
is the log likelihood specified in the MODEL
statement. The MODEL
statement specifies the log likelihood for a single observation in the data set.
If you want to model dependent data—that is, 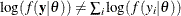 —you can use the JOINTMODEL
option in the PROC MCMC statement. See the section Modeling Joint Likelihood for more details.
—you can use the JOINTMODEL
option in the PROC MCMC statement. See the section Modeling Joint Likelihood for more details.
The statements in PROC MCMC are similar to DATA step statements; PROC MCMC evaluates every statement in order for each observation. At the beginning of the data set, the log likelihood is set to be 0. As PROC MCMC steps through the data set, it cumulatively adds the log likelihood for each observation. Statements between the BEGINNODATA and ENDNODATA statements are evaluated only at the first and the last observations. At the last observation, the log of the prior and hyperprior distributions is added to the sum of the log likelihood to obtain the log of the posterior distribution.
Calculation of the 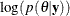 objective function involves a complete pass through the data set, making it potentially computationally expensive. If
objective function involves a complete pass through the data set, making it potentially computationally expensive. If 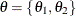 is multidimensional, you can choose to update a portion of the parameters at each iteration step by declaring them in separate
PARMS
statements (see the section Blocking of Parameters for more information). PROC MCMC updates each block of parameters while holding others constant. The objective functions
that are used in each update are the same as the log of the joint posterior density:
is multidimensional, you can choose to update a portion of the parameters at each iteration step by declaring them in separate
PARMS
statements (see the section Blocking of Parameters for more information). PROC MCMC updates each block of parameters while holding others constant. The objective functions
that are used in each update are the same as the log of the joint posterior density:
![\[ \log (p(\theta _1 | \mb{y}, \theta _2)) = \log (p(\theta _2 | \mb{y}, \theta _1)) = \log (p(\theta | \mb{y})) \]](images/statug_mcmc0120.png)
In other words, PROC MCMC does not derive the conditional distribution explicitly for each block of parameters, and it uses the full joint distribution in the Metropolis step for every block update.
Random-Effects Models
For programs that require RANDOM
statements, PROC MCMC includes the sum of the density evaluation of the random-effects parameters in the calculation of the
objective function for 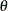 ,
,
![\[ \log (p(\theta | \bgamma , \mb{y})) = \log (\pi (\theta )) + \sum _{j=1}^ J \log (\pi (\gamma _ j | \theta )) + \sum _{i=1}^ n \log (f(y_ i|\theta , \bgamma )) \]](images/statug_mcmc0121.png)
where 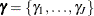 are random-effects parameters and
are random-effects parameters and 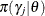 is the prior distribution of the random-effects parameters. The likelihood function can be conditional on
is the prior distribution of the random-effects parameters. The likelihood function can be conditional on 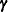 , but the prior distributions of
, but the prior distributions of 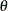 , which must be independent of
, which must be independent of 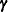 , cannot.
, cannot.
The objective function used in the Metropolis step for the random-effects parameter 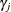 contains only the portion of the data that belong to the jth cluster:
contains only the portion of the data that belong to the jth cluster:
![\[ \log (p(\gamma _ j |\theta , \mb{y})) = \log (\pi (\gamma _ j | \theta )) + \sum _{i \in \left\{ j\mbox{th cluster} \right\} } \log (f(y_ i|\theta , \gamma _ j)) \]](images/statug_mcmc0126.png)
The calculation does not include 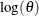 , the prior density piece, because that is a known constant. Evaluation of this objective function involves only a portion
of the data set, making it more computationally efficient. In fact, updating every random-effects parameters in a single RANDOM
statement involves only one pass through the data set.
, the prior density piece, because that is a known constant. Evaluation of this objective function involves only a portion
of the data set, making it more computationally efficient. In fact, updating every random-effects parameters in a single RANDOM
statement involves only one pass through the data set.
You can have multiple RANDOM statements in a program, which adds more pieces to the posterior calculation, such as
![\[ \log (p(\theta | \bgamma , \balpha , \mb{y})) = \log (\pi (\theta )) + \sum _{j=1}^ J \log (\pi (\gamma _ j | \theta )) + \sum _{k=1}^{K} \log (\pi (\alpha _ k | \theta )) + \sum _{i=1}^ n \log (f(y_ i|\theta , \bgamma , \balpha )) \]](images/statug_mcmc0128.png)
where 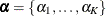 is another random effect. The random effects
is another random effect. The random effects 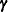 and
and  can form their own hierarchy (as in a nested model), or they can enter the program in a non-nested fashion. The objective
functions for
can form their own hierarchy (as in a nested model), or they can enter the program in a non-nested fashion. The objective
functions for 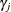 and
and 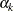 are calculated using only observations that belong to their respective clusters.
are calculated using only observations that belong to their respective clusters.
Models with Missing Values
Missing values in the response variables of the MODEL statement are treated as random variables, and they add another layer in the conditional updates in the simulation. Suppose that
![\[ \mb{y} = \left\{ \mb{y}_{\mb{obs}}, \mb{y}_{\mb{mis}} \right\} \]](images/statug_mcmc0132.png)
The response variable 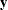 consists of
consists of 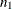 observed values
observed values 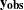 and
and 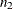 missing values
missing values 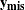 . The log of the posterior distribution is thus formed by
. The log of the posterior distribution is thus formed by
![\[ \log (p(\theta | \bgamma , \mb{y}_{\mb{mis}}, \mb{y}_{\mb{obs}})) = \log (\pi (\theta )) + \sum _{j=1}^ J \log (\pi (\gamma _ j | \theta )) + \sum _{i=1}^{n_2} \log (f(y_{\mb{mis}, i}|\theta , \bgamma )) \sum _{i=1}^{n_1} \log (f(y_{\mb{obs}, i}|\theta , \bgamma )) \]](images/statug_mcmc0137.png)
where the expression is evaluated at the drawn 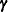 and
and 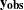 values.
values.
The conditional distribution of the random-effects parameter 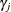 is
is
![\[ \log (p(\gamma _ j |\theta , \mb{y})) = \log (\pi (\gamma _ j | \theta )) + \sum _{i \in \left\{ j\mbox{th cluster} \right\} } \log (f(y_ i|\theta , \gamma _ j)) \]](images/statug_mcmc0126.png)
where the 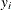 are either the observed or the imputed values of the response variable.
are either the observed or the imputed values of the response variable.
The missing values are usually sampled directly from the sampling distribution and do not require the Metropolis sampler.
When a response variable takes on a GENERAL
function, the objective function is simply the likelihood function: 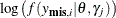 .
.