The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
Example 73.9 Multivariate Normal Random-Effects Model
Gelfand et al. (1990) use a multivariate normal hierarchical model to estimate growth regression coefficients for the growth of 30 young rats
in a control group over a period of 5 weeks. The following statements create a SAS data set with measurements of Weight, Age (in days), and Subject.
title 'Multivariate Normal Random-Effects Model'; data rats; array days[5] (8 15 22 29 36); input weight @@; subject = ceil(_n_/5); index = mod(_n_-1, 5) + 1; age = days[index]; drop index days:; datalines; 151 199 246 283 320 145 199 249 293 354 147 214 263 312 328 155 200 237 272 297 135 188 230 280 323 159 210 252 298 331 141 189 231 275 305 159 201 248 297 338 177 236 285 350 376 134 182 220 260 296 160 208 261 313 352 143 188 220 273 314 154 200 244 289 325 171 221 270 326 358 163 216 242 281 312 160 207 248 288 324 142 187 234 280 316 156 203 243 283 317 157 212 259 307 336 152 203 246 286 321 154 205 253 298 334 139 190 225 267 302 146 191 229 272 302 157 211 250 285 323 132 185 237 286 331 160 207 257 303 345 169 216 261 295 333 157 205 248 289 316 137 180 219 258 291 153 200 244 286 324 ;
The model assumes normal measurement errors,
![\[ \mbox{Weight}_{ij} \sim \mbox{normal} \left( \alpha _ i + \beta _ i \mbox{Age}_{ij}, \sigma ^2 \right), ~ ~ ~ i = 1, \ldots , 30; j = 1, \ldots , 5 \]](images/statug_mcmc0691.png)
where i indexes rat (Subject variable), j indexes the time period, Weight and Age
and Age denote the weight and age of the ith rat in week j, and
denote the weight and age of the ith rat in week j, and 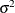 is the variance in the normal likelihood. The individual intercept and slope coefficients are modeled as the following:
is the variance in the normal likelihood. The individual intercept and slope coefficients are modeled as the following:
![\[ \bm {\theta }_ i = \left(\begin{matrix} \alpha _ i \\ \beta _ i \end{matrix} \right) \sim \mbox{MVN} \left( \bm {\theta }_ c = \begin{pmatrix} \alpha _ c \\ \beta _ c \end{pmatrix}, \bm {\Sigma }_ c \right), ~ ~ ~ i = 1, \ldots , 30 \]](images/statug_mcmc0693.png)
You can use the following independent prior distributions on 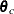 ,
, 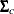 , and
, and 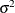 :
:
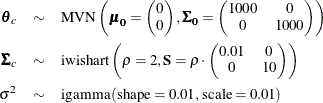
The following statements fit this multivariate normal random-effects model:
proc mcmc data=rats nmc=10000 outpost=postout
seed=17 init=random;
ods select Parameters REParameters PostSumInt;
array theta[2] alpha beta;
array theta_c[2];
array Sig_c[2,2];
array mu0[2] (0 0);
array Sig0[2,2] (1000 0 0 1000);
array S[2,2] (0.02 0 0 20);
parms theta_c Sig_c {121 0 0 0.26} var_y;
prior theta_c ~ mvn(mu0, Sig0);
prior Sig_c ~ iwish(2, S);
prior var_y ~ igamma(0.01, scale=0.01);
random theta ~ mvn(theta_c, Sig_c) subject=subject
monitor=(alpha_9 beta_9 alpha_25 beta_25);
mu = alpha + beta * age;
model weight ~ normal(mu, var=var_y);
run;
The ODS SELECT statement displays information about model parameters, random-effects parameters, and the posterior summary
statistics. The ARRAY
statements allocate memory space for the multivariate parameters and hyperparameters in the model. The parameters are 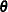 (
(theta where the variable name of each element is alpha or beta), 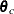 (
(theta_c), and 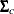 (
(Sig_c). The hyperparameters are 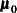 (
(mu0), 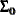 (
(Sig0), and 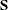 (
(S). The multivariate hyperparameters are assigned with constant values using parentheses  .
.
The PARMS
statement declares model parameters and assigns initial values to Sig_c using braces 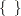 . The PRIOR
statements specify the prior distributions. The RANDOM
statement defines an array random effect
. The PRIOR
statements specify the prior distributions. The RANDOM
statement defines an array random effect theta and specifies a multivariate normal prior distribution. The SUBJECT=
option indicates cluster membership for each of the random-effects parameter. The MONITOR=
option monitors the individual intercept and slope coefficients of subjects 9 and 25.
You can use the following syntax in the RANDOM statement to monitor all parameters in an array random effect:
monitor=(theta)
This would produce posterior summary statistics on 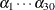 and
and 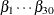 .
.
The following syntax monitors all 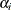 parameters:
parameters:
monitor=(alpha)
If you did not name elements of theta to be alpha and beta, the SAS System creates variable names automatically in a consecutive fashion, as in theta1 and theta2.
Output 73.9.1: Parameter and Random-Effects Parameter Information Table
| Multivariate Normal Random-Effects Model |
| Parameters | |||||
|---|---|---|---|---|---|
| Block | Parameter | Array Index |
Sampling Method |
Initial Value |
Prior Distribution |
| 1 | theta_c1 | Conjugate | -4.5834 | MVNormal(mu0, Sig0) | |
| theta_c2 | 5.7930 | ||||
| 2 | Sig_c1 | [1,1] | Conjugate | 121.0 | iWishart(2, S) |
| Sig_c2 | [1,2] | 0 | |||
| Sig_c3 | [2,1] | 0 | |||
| Sig_c4 | [2,2] | 0.2600 | |||
| 3 | var_y | Conjugate | 2806714 | igamma(0.01, scale=0.01) | |
Output 73.9.1 displays the parameter and random-effects parameter information tables. The Array Index column in "Parameters" table shows the index reference of the elements in the array parameter Sig_c. The total number of subjects in the study is 30.
Output 73.9.2: Multivariate Normal Random-Effects Model
| Multivariate Normal Random-Effects Model |
| Posterior Summaries and Intervals | |||||
|---|---|---|---|---|---|
| Parameter | N | Mean | Standard Deviation |
95% HPD Interval | |
| theta_c1 | 10000 | 106.1 | 2.2486 | 101.7 | 110.6 |
| theta_c2 | 10000 | 6.1975 | 0.1988 | 5.8058 | 6.5815 |
| Sig_c1 | 10000 | 110.8 | 45.9169 | 37.9670 | 203.8 |
| Sig_c2 | 10000 | -1.4267 | 2.3320 | -6.2878 | 2.7756 |
| Sig_c3 | 10000 | -1.4267 | 2.3320 | -6.2878 | 2.7756 |
| Sig_c4 | 10000 | 1.0591 | 0.2979 | 0.5549 | 1.6538 |
| var_y | 10000 | 37.6855 | 5.9591 | 27.0943 | 49.4449 |
| alpha_9 | 10000 | 119.4 | 5.6756 | 108.1 | 130.5 |
| beta_9 | 10000 | 7.4670 | 0.2382 | 7.0146 | 7.9278 |
| alpha_25 | 10000 | 86.5673 | 6.3694 | 74.4247 | 99.9007 |
| beta_25 | 10000 | 6.7804 | 0.2612 | 6.2529 | 7.2906 |
Output 73.9.2 displays posterior summary statistics for model parameters and the random-effects parameters for subjects 9 and 25. You can
see that there is a substantial difference in the intercepts and growth rates between the two rats.
A seemingly confusing message might occur if a symbol name matches an internally generated variable name for elements of an
array. For example, if, instead of using the symbol var_y in the SAS program for the model variance 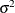 , you used
, you used s2, the SAS System produces the following error message:
ERROR: The initial value 0 for the parameter S2 is outside of the prior
distribution support set.
This is confusing because the program does not assign an initial value for the parameter s2 in the PARMS
statement, and you might expect that PROC MCMC would not generate an invalid initial value. The confusion is caused by the
ARRAY
statement that defines the array variable S:
array S[2,2] (0.02 0 0 20);
Elements of S are automatically given names s1–s4. PROC MCMC interprets s2 as an element in S that was given a value of 0, hence producing this error message.