The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
Example 73.4 Logistic Regression Model with Jeffreys’ Prior
A controlled experiment was run to study the effect of the rate and volume of air inspired on a transient reflex vasoconstriction
in the skin of the fingers. Thirty-nine tests under various combinations of rate and volume of air inspired were obtained
(Finney 1947). The result of each test is whether or not vasoconstriction occurred. Pregibon (1981) uses this set of data to illustrate the diagnostic measures he proposes for detecting influential observations and to quantify
their effects on various aspects of the maximum likelihood fit. The following statements create the data set Vaso:
title 'Logistic Regression Model with Jeffreys Prior'; data vaso; input vol rate resp @@; lvol = log(vol); lrate = log(rate); ind = _n_; cnst = 1; datalines; 3.7 0.825 1 3.5 1.09 1 1.25 2.5 1 0.75 1.5 1 0.8 3.2 1 0.7 3.5 1 0.6 0.75 0 1.1 1.7 0 0.9 0.75 0 0.9 0.45 0 0.8 0.57 0 0.55 2.75 0 0.6 3.0 0 1.4 2.33 1 0.75 3.75 1 2.3 1.64 1 3.2 1.6 1 0.85 1.415 1 1.7 1.06 0 1.8 1.8 1 0.4 2.0 0 0.95 1.36 0 1.35 1.35 0 1.5 1.36 0 1.6 1.78 1 0.6 1.5 0 1.8 1.5 1 0.95 1.9 0 1.9 0.95 1 1.6 0.4 0 2.7 0.75 1 2.35 0.03 0 1.1 1.83 0 1.1 2.2 1 1.2 2.0 1 0.8 3.33 1 0.95 1.9 0 0.75 1.9 0 1.3 1.625 1 ;
The variable resp represents the outcome of a test. The variable lvol represents the log of the volume of air intake, and the variable lrate represents the log of the rate of air intake. You can model the data by using logistic regression. You can model the response
with a binary likelihood:
![\[ \mbox{resp}_ i \sim \mbox{binary}(p_ i) \]](images/statug_mcmc0647.png)
with
![\[ p_ i = \frac{1}{1+ \exp (- (\beta _0 + \beta _1 \mbox{lvol}_ i + \beta _2 \mbox{lrate}_ i))} \]](images/statug_mcmc0648.png)
Let 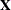 be the design matrix in the regression. Jeffreys’ prior for this model is
be the design matrix in the regression. Jeffreys’ prior for this model is
![\[ p(\bbeta ) \propto |\bX ’ \bM \bX |^{1/2} \]](images/statug_mcmc0650.png)
where 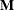 is a 39 by 39 matrix with off-diagonal elements being 0 and diagonal elements being
is a 39 by 39 matrix with off-diagonal elements being 0 and diagonal elements being 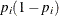 . For details on Jeffreys’ prior, see Jeffreys’ Prior in Chapter 7: Introduction to Bayesian Analysis Procedures. You can use a number of matrix functions, such as the determinant function, in PROC MCMC to construct Jeffreys’ prior. The
following statements illustrate how to fit a logistic regression with Jeffreys’ prior:
. For details on Jeffreys’ prior, see Jeffreys’ Prior in Chapter 7: Introduction to Bayesian Analysis Procedures. You can use a number of matrix functions, such as the determinant function, in PROC MCMC to construct Jeffreys’ prior. The
following statements illustrate how to fit a logistic regression with Jeffreys’ prior:
%let n = 39;
proc mcmc data=vaso nmc=10000 outpost=mcmcout seed=17;
ods select PostSumInt;
array beta[3] beta0 beta1 beta2;
array m[&n, &n];
array x[1] / nosymbols;
array xt[3, &n];
array xtm[3, &n];
array xmx[3, 3];
array p[&n];
parms beta0 1 beta1 1 beta2 1;
begincnst;
if (ind eq 1) then do;
rc = read_array("vaso", x, "cnst", "lvol", "lrate");
call transpose(x, xt);
call zeromatrix(m);
end;
endcnst;
beginnodata;
call mult(x, beta, p); /* p = x * beta */
do i = 1 to &n;
p[i] = 1 / (1 + exp(-p[i])); /* p[i] = 1/(1+exp(-x*beta)) */
m[i,i] = p[i] * (1-p[i]);
end;
call mult (xt, m, xtm); /* xtm = xt * m */
call mult (xtm, x, xmx); /* xmx = xtm * x */
call det (xmx, lp); /* lp = det(xmx) */
lp = 0.5 * log(lp); /* lp = -0.5 * log(lp) */
prior beta: ~ general(lp);
endnodata;
model resp ~ bern(p[ind]);
run;
The first ARRAY
statement defines an array beta with three elements: beta0, beta1, and beta2. The subsequent statements define arrays that are used in the construction of Jeffreys’ prior. These include m (the 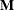 matrix),
matrix), x (the design matrix), xt (the transpose of x), and some additional work spaces.
The explanatory variables lvol and lrate are saved in the array x in the BEGINCNST
and ENDCNST
statements. See BEGINCNST/ENDCNST Statement for details. After all the variables are read into x, you transpose the x matrix and store it to xt. The ZEROMATRIX function call assigns all elements in matrix m the value zero. To avoid redundant calculation, it is best to perform these calculations as the last observation of the data
set is processed—that is, when ind is 39.
You calculate Jeffreys’ prior in the BEGINNODATA
and ENDNODATA
statements. The probability vector p is the product of the design matrix x and parameter vector beta. The diagonal elements in the matrix m are 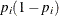 . The expression
. The expression lp is the logarithm of Jeffreys’ prior. The PRIOR
statement assigns lp as the prior for the 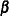 regression coefficients. The MODEL
statement assigns a binary likelihood to
regression coefficients. The MODEL
statement assigns a binary likelihood to resp, with probability p[ind]. The p array is calculated earlier using the matrix function MULT. You use the ind variable to pick out the right probability value for each resp.
Posterior summary statistics are displayed in Output 73.4.1.
Output 73.4.1: PROC MCMC Results, Jeffreys’ prior
You can also use PROC GENMOD to fit the same model by using the following statements:
proc genmod data=vaso descending; ods select PostSummaries PostIntervals; model resp = lvol lrate / d=bin link=logit; bayes seed=17 coeffprior=jeffreys nmc=20000 thin=2; run;
The MODEL statement indicates that resp is the response variable and lvol and lrate are the covariates. The options in the MODEL statement specify a binary likelihood and a logit link function. The BAYES statement
requests Bayesian capability. The SEED=, NMC=, and THIN= arguments work in the same way as in PROC MCMC. The COEFFPRIOR=JEFFREYS
option requests Jeffreys’ prior in this analysis.
The PROC GENMOD statements produce Output 73.4.2, with estimates very similar to those reported in Output 73.4.1. Note that you should not expect to see identical output from PROC GENMOD and PROC MCMC, even with the simulation setup and identical random number seed. The two procedures use different sampling algorithms. PROC GENMOD uses the adaptive rejection metropolis algorithm (ARMS) (Gilks and Wild 1992; Gilks 2003) while PROC MCMC uses a random walk Metropolis algorithm. The asymptotic answers, which means that you let both procedures run an very long time, would be the same as they both generate samples from the same posterior distribution.
Output 73.4.2: PROC GENMOD Results
| Logistic Regression Model with Jeffreys Prior |
| Posterior Summaries | ||||||
|---|---|---|---|---|---|---|
| Parameter | N | Mean | Standard Deviation |
Percentiles | ||
| 25% | 50% | 75% | ||||
| Intercept | 10000 | -2.8773 | 1.3213 | -3.6821 | -2.7326 | -1.9097 |
| lvol | 10000 | 5.2059 | 1.8707 | 3.8535 | 4.9574 | 6.3337 |
| lrate | 10000 | 4.5525 | 1.8140 | 3.2281 | 4.3722 | 5.6643 |