The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
Example 73.8 Nonlinear Poisson Regression Multilevel Random-Effects Model
This example uses the pump failure data of Gaver and O’Muircheartaigh (1987) to illustrate how to fit a multilevel random-effects model with PROC MCMC. The number of failures and the time of operation are recorded for 10 pumps. Each of the pumps is classified into one of two groups that correspond to either continuous or intermittent operation. The following statements generate the data set:
title 'Nonlinear Poisson Regression Random-Effects Model'; data pump; input y t group @@; pump = _n_; logtstd = log(t) - 2.4564900; datalines; 5 94.320 1 1 15.720 2 5 62.880 1 14 125.760 1 3 5.240 2 19 31.440 1 1 1.048 2 1 1.048 2 4 2.096 2 22 10.480 2 ;
Each row denotes data for a single pump, and the variable logtstd contains the centered operation times. Letting 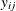 denote the number of failures for the jth pump in the ith group, Draper (1996) considers the following hierarchical model for these data, where the data set variable
denote the number of failures for the jth pump in the ith group, Draper (1996) considers the following hierarchical model for these data, where the data set variable logtstd is 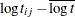 :
:
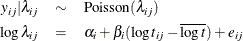
This model specifies different intercepts and slopes for each group (i = 1, 2), and the random effect 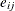 is a mechanism for accounting for overdispersion. You can use noninformative priors on the parameters
is a mechanism for accounting for overdispersion. You can use noninformative priors on the parameters 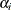 ,
, 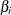 , and
, and 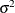 , and a normal prior on
, and a normal prior on 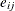 ,
,
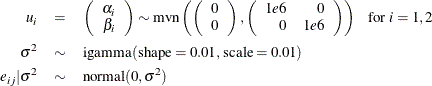
where 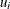 is a multidimensional random effect. The following statements fit such a random-effects model:
is a multidimensional random effect. The following statements fit such a random-effects model:
ods graphics on; proc mcmc data=pump outpost=postout seed=248601 nmc=10000 plots=trace stats=none diag=none; ods select tracepanel; array u[2] alpha beta; array mu[2] (0 0); parms s2; prior s2 ~ igamma(0.01, scale=0.01); random u ~ MVNAR(mu, sd=1e6, rho=0) subject=group monitor=(u); random e ~ normal(0, var=s2) subject=pump monitor=(random(1)); w = alpha + beta * logtstd; lambda = exp(w+e); model y ~ poisson(lambda); run;
The PROC MCMC statement specifies the input data set (Pump), the output data set (Postout), a seed for the random number generator, and a simulation sample size of 10,000. The program requests that only trace plots
be produced, disabling all posterior calculations and convergence diagnostics tests. The ODS SELECT statement displays the
trace plots, which are the primary focus.
The first ARRAY
statement declares an array u of size 2 and names the elements alpha and beta. The array u stores the random-effects parameters alpha and beta. The next ARRAY
statement defines the mean of the multivariate normal prior on u.
The PARMS
statement declares the only model parameter here, the variance s2 in the prior distribution for the random effect 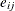 . The PRIOR
statement specifies an inverse-gamma prior on the variance. The first RANDOM
statement specifies a multivariate normal prior on
. The PRIOR
statement specifies an inverse-gamma prior on the variance. The first RANDOM
statement specifies a multivariate normal prior on u. The MVNAR
distribution is a multivariate normal distribution with a first-order autoregressive covariance. When the argument rho is set to 0, this distribution simplifies to a multivariate normal distribution with a shared variance. The RANDOM
statement also indicates the group variable as its subject index and monitors all elements u. The second RANDOM
statement specifies a normal prior on the effect e, where the subject index variable is pump. The MONITOR=
option requests that PROC MCMC randomly choose one of the 10 e random-effects parameters to monitor.
Next, programming statements construct the mean of the Poisson likelihood, and the MODEL statement specifies the likelihood function for each observation.
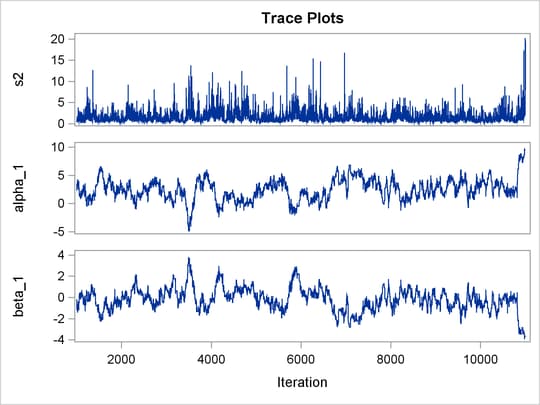
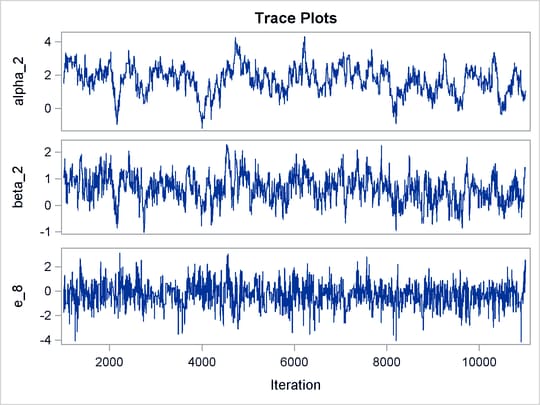
Output 73.8.1 shows trace plots for 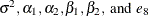 . You can see that the chains are mixing poorly.
. You can see that the chains are mixing poorly.
Output 73.8.1: Trace Plots of 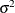 ,
,  ,
, 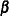 , and
, and 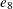 without Hierarchical Centering
without Hierarchical Centering
To improve mixing, you can repeat the same analysis by using a hierarchical centering technique, where instead of using a
normal prior centered at 0 on 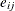 , you center the random effects on the group means:
, you center the random effects on the group means:
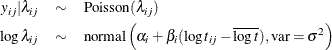
The following statements illustrate how to fit a multilevel hierarchical centering random-effects model:
proc mcmc data=pump outpost=postout_c seed=248601 nmc=10000 plots=trace diag=none; ods select tracepanel postsumint; array u[2] alpha beta; array mu[2] (0 0); parms s2 1; prior s2 ~ igamma(0.01, scale=0.01); random u ~ MVNAR(mu, sd=1e6, rho=0) subject=group monitor=(u); w = alpha + beta * logtstd; random llambda ~ normal(w, var = s2) subject=pump monitor=(random(1)); lambda = exp(llambda); model y ~ poisson(lambda); run;
The difference between these statements and the previous statements on is that these statements have the variable w as the prior mean of the random effect llambda. The symbol lambda is the exponential of the corresponding 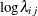 (
(llambda), and the MODEL
statement assigns the response variable y a Poisson likelihood with a mean parameter lambda, the same way it did in the previous statements.
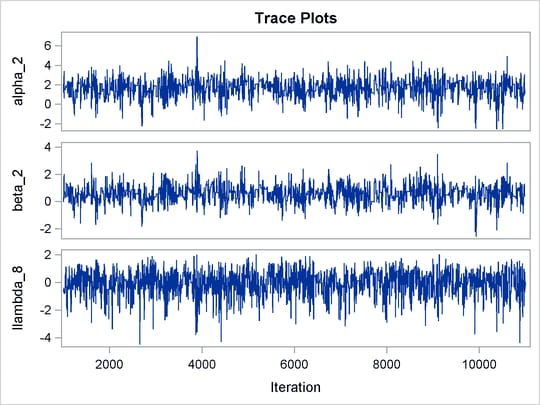
The trace plots of the monitored parameters are shown in Output 73.8.2. The mixing is significantly improved over the previous model. The posterior summary and interval statistics tables are shown in Output 73.8.3.
Output 73.8.2: Trace Plots of 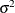 ,
,  , and
, and 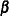 with Hierarchical Centering
with Hierarchical Centering
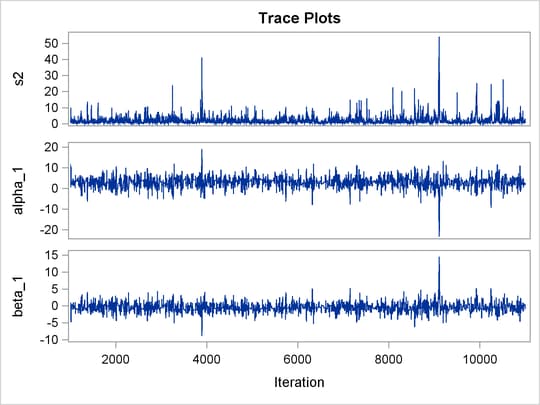
Output 73.8.3: Posterior Summary Statistics
| Nonlinear Poisson Regression Random-Effects Model |
| Posterior Summaries and Intervals | |||||
|---|---|---|---|---|---|
| Parameter | N | Mean | Standard Deviation |
95% HPD Interval | |
| s2 | 10000 | 1.7606 | 2.2022 | 0.1039 | 4.7631 |
| alpha_1 | 10000 | 2.9286 | 2.4247 | -1.9416 | 7.4115 |
| beta_1 | 10000 | -0.4018 | 1.3110 | -2.9323 | 2.0623 |
| alpha_2 | 10000 | 1.6105 | 0.8801 | -0.0436 | 3.2985 |
| beta_2 | 10000 | 0.5652 | 0.5804 | -0.5469 | 1.7381 |
| llambda_8 | 10000 | -0.0560 | 0.8395 | -1.6933 | 1.4612 |
You can generate a caterpillar plot (Output 73.8.4) of the random-effects parameters by calling the %CATER macro:
%CATER(data=postout_c, var=llambda_:); ods graphics off;
Varying llambda indicates nonconstant dispersion in the Poisson model.
Output 73.8.4: Caterpillar Plots of the Random-Effects Parameters