The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
Example 73.12 Change Point Models
Consider the data set from Bacon and Watts (1971), where 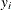 is the logarithm of the height of the stagnant surface layer and the covariate
is the logarithm of the height of the stagnant surface layer and the covariate 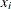 is the logarithm of the flow rate of water. The following statements create the data set:
is the logarithm of the flow rate of water. The following statements create the data set:
title 'Change Point Model'; data stagnant; input y x @@; ind = _n_; datalines; 1.12 -1.39 1.12 -1.39 0.99 -1.08 1.03 -1.08 0.92 -0.94 0.90 -0.80 0.81 -0.63 0.83 -0.63 0.65 -0.25 0.67 -0.25 0.60 -0.12 0.59 -0.12 0.51 0.01 0.44 0.11 0.43 0.11 0.43 0.11 0.33 0.25 0.30 0.25 0.25 0.34 0.24 0.34 0.13 0.44 -0.01 0.59 -0.13 0.70 -0.14 0.70 -0.30 0.85 -0.33 0.85 -0.46 0.99 -0.43 0.99 -0.65 1.19 ;
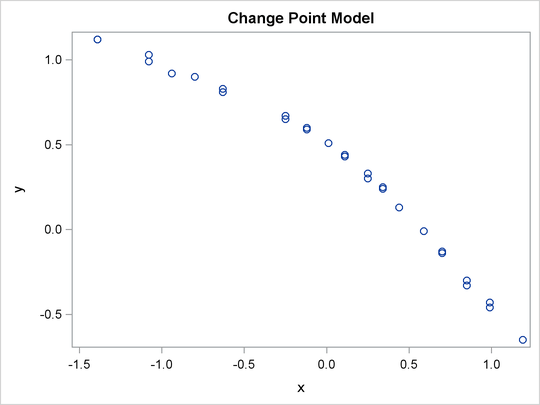
A scatter plot (Output 73.12.1) shows the presence of a nonconstant slope in the data. This suggests a change point regression model (Carlin, Gelfand, and Smith 1992). The following statements generate the scatter plot in Output 73.12.1:
proc sgplot data=stagnant; scatter x=x y=y; run;
Output 73.12.1: Scatter Plot of the Stagnant Data Set
Let the change point be cp. Following formulation by Spiegelhalter et al. (1996b), the regression model is as follows:
![\[ y_ i \sim \left\{ \begin{array}{ll} \mbox{normal}(\alpha + \beta _1 (x_ i - {\Variable{cp}}), \sigma ^2) & \mbox{if } x_ i < {\Variable{cp}} \\ \mbox{normal}(\alpha + \beta _2 (x_ i - {\Variable{cp}}), \sigma ^2) & \mbox{if } x_ i >= {\Variable{cp}} \end{array} \right. \]](images/statug_mcmc0723.png)
You might consider the following diffuse prior distributions:

The following statements generate Output 73.12.2:
proc mcmc data=stagnant outpost=postout seed=24860 ntu=1000
nmc=20000;
ods select PostSumInt;
ods output PostSumInt=ds;
array beta[2];
parms alpha cp beta1 beta2;
parms s2;
prior cp ~ unif(-1.3, 1.1);
prior s2 ~ uniform(0, 5);
prior alpha beta: ~ normal(0, v = 1e6);
j = 1 + (x >= cp);
mu = alpha + beta[j] * (x - cp);
model y ~ normal(mu, var=s2);
run;
The PROC MCMC statement specifies the input data set (Stagnant), the output data set (Postout), a random number seed, a tuning sample of 1000, and an MCMC sample of 20000. The ODS SELECT statement displays only the
summary statistics table. The ODS OUTPUT statement saves the summary statistics table to the data set Ds.
The ARRAY
statement allocates an array of size 2 for the beta parameters. You can use beta1 and beta2 as parameter names without allocating an array, but having the array makes it easier to construct the likelihood function.
The two PARMS
statements put the five model parameters in two blocks. The three PRIOR
statements specify the prior distributions for these parameters.
The symbol j indicates the segment component of the regression. When x is less than the change point, (x >= cp) returns 0 and j is assigned the value 1; if x is greater than or equal to the change point, (x >= cp) returns 1 and j is 2. The symbol mu is the mean for the jth segment, and beta[j] changes between the two regression coefficients depending on the segment component. The MODEL
statement assigns the normal model to the response variable y.
Posterior summary statistics are shown in Output 73.12.2.
Output 73.12.2: MCMC Estimates of the Change Point Regression Model
| Change Point Model |
| Posterior Summaries and Intervals | |||||
|---|---|---|---|---|---|
| Parameter | N | Mean | Standard Deviation |
95% HPD Interval | |
| alpha | 20000 | 0.5349 | 0.0249 | 0.4843 | 0.5813 |
| cp | 20000 | 0.0283 | 0.0314 | -0.0353 | 0.0846 |
| beta1 | 20000 | -0.4200 | 0.0146 | -0.4482 | -0.3911 |
| beta2 | 20000 | -1.0136 | 0.0167 | -1.0476 | -0.9817 |
| s2 | 20000 | 0.000451 | 0.000145 | 0.000220 | 0.000735 |
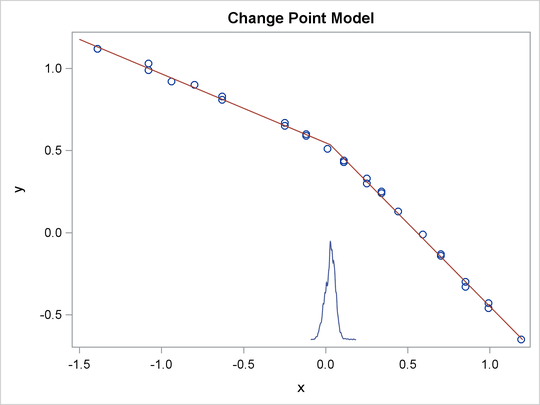
You can use PROC SGPLOT to visualize the model fit. Output 73.12.3 shows the fitted regression lines over the original data. In addition, on the bottom of the plot is the kernel density of
the posterior marginal distribution of cp, the change point. The kernel density plot shows the relative variability of the posterior distribution on the data plot.
You can use the following statements to create the plot:
data _null_; set ds; call symputx(parameter, mean); run; data b; missing A; input x1 @@; if x1 eq .A then x1 = &cp; if _n_ <= 2 then y1 = &alpha + &beta1 * (x1 - &cp); else y1 = &alpha + &beta2 * (x1 - &cp); datalines; -1.5 A 1.2 ; proc kde data=postout; univar cp / out=m1 (drop=count); run; data m1; set m1; density = (density / 25) - 0.653; run; data all; set stagnant b m1; run; proc sgplot data=all noautolegend; scatter x=x y=y; series x=x1 y=y1 / lineattrs = graphdata2; series x=value y=density / lineattrs = graphdata1; run;
The macro variables &alpha, &beta1, &beta2, and &cp store the posterior mean estimates from the data set Ds. The data set b contains three predicted values, at the minimum and maximum values of x and the estimated change point &cp. These input values give you fitted values from the regression model. Data set M1 contains the kernel density estimates of the parameter cp. The density is scaled down so the curve would fit in the plot. Finally, you use PROC SGPLOT to overlay the scatter plot,
regression line and kernel density plots in the same graph.
Output 73.12.3: Estimated Fit to the Stagnant Data Set