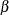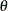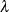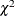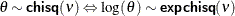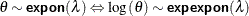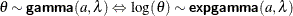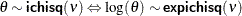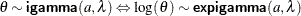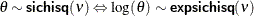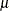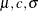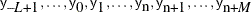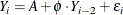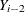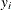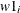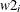The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
MODEL Statement
-
MODEL dependent-variable-list ~distribution <options>;
The MODEL statement specifies the conditional distribution of the data given the parameters (the likelihood function). You specify a single dependent variable or a list of dependent variables, a tilde ~, and then a distribution with its arguments. The dependent variables can be variables from the input data set or functions of the symbols in the program. You must specify the dependent variables unless you use the GENERAL function or the DGENERAL function (see the section Specifying a New Distribution for more details).
The MODEL statement assumes that the observations are independent of each other, conditional on the model parameters. If you
want to model dependent data—that is, 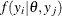 for
for 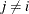 —you can use the JOINTMODEL
option in the PROC MCMC statement. See the section Modeling Joint Likelihood for more details. By default, the log-likelihood value is the sum of the individual log-likelihood value for each observation.
—you can use the JOINTMODEL
option in the PROC MCMC statement. See the section Modeling Joint Likelihood for more details. By default, the log-likelihood value is the sum of the individual log-likelihood value for each observation.
You can specify multiple MODEL statements. You can define likelihood functions that are independent of each other. For example,
in the following statements, the dependent variables y1 and y2 are independent of each other:
model y1 ~ normal(alpha, var=s21); model y2 ~ normal(beta, var=s22);
Alternatively, you can use marginal and conditional distributions to define a joint log-likelihood function for multiple dependent
variables. For example, the following statements jointly define a distribution over 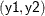 . They specify a marginal distribution for the dependent variable
. They specify a marginal distribution for the dependent variable y1 and a conditional distribution for the dependent variable y2:
model y1 ~ normal(alpha, var=s21); model y2 ~ normal(beta * y1, var=s22);
Every program must have at least one MODEL statement. If you want to run a Monte Carlo simulation that does not require a response variable, use the GENERAL function in the MODEL statement:
model general(0);
PROC MCMC interprets the statement as a flat likelihood function with a constant log-likelihood value of 0.
PROC MCMC is a programming language that is similar to the DATA step, and the order of statement evaluation is important. For example, the MODEL statement must come after any SAS programming statements that define or modify arguments used in the construction of the log likelihood. In PROC MCMC, a symbol can be defined multiple times and used at different places. Using an expression out of order produces erroneous results that can also be hard to detect.
Do not embed the MODEL statement within programming statements. For example, suppose you have three response variables, y1, y2, and y3, and want to model each with a normal distribution. The following statements lead to erroneous output:
array Y[3] y1 y2 y3; do i = 1 to 3; model y[i] ~ normal(mu, sd=s); end;
Instead, you should do one of the following.
-
Use separate MODEL statements:
model y1 ~ normal(mu, sd=s); model y2 ~ normal(mu, sd=s); model y3 ~ normal(mu, sd=s);
-
Use the GENERAL function to construct a joint distribution of the three dependent variables and use a single MODEL statement to specify the log-likelihood function:
llike = logpdf("normal", y1, mu, s) + logpdf("normal", y2, mu, s) + logpdf("normal", y3, mu, s); model y1 y2 y3 ~ general(llike);See the section Specifying a New Distribution for more information about how to use the GENERAL function to specify an arbitrary distribution.
Missing data are allowed in the response variables; the MODEL statement augments missing data automatically. (In releases before SAS/STAT 12.1, observations with missing values were discarded prior to analysis and PROC MCMC did not attempt to model these values.) In each iteration, PROC MCMC samples missing values from their posterior distributions and incorporates them as part of the simulation. PROC MCMC creates one variable for each missing response value. There are two ways to create the missing value variable names; see the NAMESUFFIX= option for the naming convention of the variables.
Distributions in MODEL Statement
Standard distributions that the MODEL statement supports are listed in the Table 73.2 (univariate) and Table 73.3 (multivariate). See the section Standard Distributions for density specifications. You can also specify all distributions except the multinomial distribution in the PRIOR and HYPERPRIOR statements. The RANDOM statement supports only a subset of the distributions (see Table 73.4).
PROC MCMC allows some distributions to be parameterized in multiple ways. For example, you can specify a normal distribution with a variance, standard deviation, or precision parameter. For distributions that have different parameterizations, you must specify an option to clearly name the ambiguous parameter. For example, in the normal distribution, you must indicate whether the second argument represents variance, standard deviation, or precision.
All univariate distributions, with the exception of binary and uniform, can have the optional LOWER= and UPPER= arguments, which specify a truncated density. See the section Truncation and Censoring for more details. Truncation is not supported for multivariate distributions.
Table 73.2: Univariate Distributions
|
Distribution Name |
Definition |
|---|---|
|
beta
(<a=> |
Beta distribution with shape parameters |
|
binary (<prob|p=> p) |
Binary (Bernoulli) distribution with probability of success p. You can use the alias bern for this distribution. |
|
binomial (<n=> n, <prob|p=> p) |
Binomial distribution with count n and probability of success p |
|
cauchy
(<location|loc|l=> |
Cauchy distribution with location |
|
chisq
(<df=> |
|
|
dgeneral (ll) |
General log-likelihood function that you construct using SAS programming statements for single or multiple discrete parameters. Also see the function general. The name dlogden is an alias for this function. |
|
expchisq
(<df=> |
Log transformation of a |
|
Log transformation of an exponential distribution with scale or inverse-scale parameter |
|
|
expGamma
(<shape|sp=> a, scale|s= |
Log transformation of a gamma distribution with shape a and scale or inverse-scale |
|
expichisq
(<df=> |
Log transformation of an inverse |
|
expiGamma
(<shape|sp=> a, scale|s= |
Log transformation of an inverse-gamma distribution with shape a and scale or inverse-scale |
|
expsichisq
(<df=> |
Log transformation of a scaled inverse |
|
Exponential distribution with scale or inverse-scale parameter |
|
|
gamma
(<shape|sp=> a, scale|s= |
Gamma distribution with shape a and scale or inverse-scale |
|
geo (<prob|p=> p) |
Geometric distribution with probability p |
|
general (ll) |
General log-likelihood function that you construct using SAS programming statements for a single or multiple continuous parameters. The argument ll is an expression for the log of the distribution. If there are multiple variables specified before the tilde in a MODEL, PRIOR, or HYPERPRIOR statement, ll is interpreted as the log of the joint distribution for these variables. Note that in the MODEL statement, the response variable specified before the tilde is just a place holder and is of no consequence; the variable must have appeared in the construction of ll in the programming statements. general(constant) is equivalent to a uniform distribution on the real line. You can use the alias logden for this distribution. |
|
ichisq
(<df=> |
Inverse |
|
igamma
(<shape|sp=> a, scale|s= |
Inverse-gamma distribution with shape a and scale or inverse-scale |
|
laplace
(<location|loc|l=> |
Laplace distribution with location |
|
logistic (<location|loc|l=> a, <scale|s=> b) |
Logistic distribution with location a and scale b |
|
lognormal
(<mean|m=> |
Log-normal distribution with mean |
|
negbin (<n=> n, <prob|p=> p) |
Negative binomial distribution with count n and probability of success p. You can use the alias nb for this distribution. |
|
normal
(<mean|m=> |
Normal (Gaussian) distribution with mean |
|
pareto (<shape|sp=> a, <scale|s=> b) |
Pareto distribution with shape a and scale b |
|
poisson
(<mean|m=> |
Poisson distribution with mean |
|
sichisq
(<df=> |
Scaled inverse |
|
t
(<mean|m=> |
T distribution with mean |
|
table
(<p=> p) |
Table (categorical) distribution with probability vector p. You can also use the alias cat for this distribution. |
|
uniform (<left|l=> a, <right|r=> b) |
Uniform distribution with range a and b. You can use the alias unif for this distribution. |
|
wald
(<mean|m=> |
Wald distribution with mean parameter |
|
weibull
( |
Weibull distribution with location (threshold) parameter |
Table 73.3: Multivariate Distributions
|
Distribution Name |
Definition |
|---|---|
|
dirichlet
(<alpha=> |
Dirichlet distribution with parameter vector |
|
iwish
(<df=> |
Inverse Wishart distribution with |
|
multinom (<p=>p) |
Multinomial distribution with probability vector p |
|
mvn
(<mu=> |
Multivariate normal distribution with mean vector |
|
MVNAR
(<mu=> |
Multivariate normal distribution with mean vector |
Options for the MODEL Statement
The options in the MODEL statement apply when there are missing values in the response variable, or in the case of the ICOND= option, when there are lag or lead variables for the response variable. You can specify the following options.