The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
Example 73.22 One-Compartment Model with Pharmacokinetic Data
A popular application of nonlinear mixed models is in the field of pharmacokinetics, which studies how a drug disperses through a living individual. This example considers the theophylline data from Pinheiro and Bates (1995). Serum concentrations of the drug theophylline, which is used to treat respiratory diseases, are measured in 12 subjects over a 25-hour period after oral administration. The data are as follows.
data theoph; input subject time conc dose; datalines; 1 0.00 0.74 4.02 1 0.25 2.84 4.02 1 0.57 6.57 4.02 1 1.12 10.50 4.02 1 2.02 9.66 4.02 1 3.82 8.58 4.02 1 5.10 8.36 4.02 1 7.03 7.47 4.02 ... more lines ... 12 24.15 1.17 5.30 ;
A commonly used one-compartment model is based on the differential equations
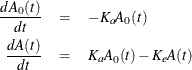
where 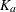 is the absorption rate and
is the absorption rate and 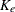 is the elimination rate.
is the elimination rate.
The initial values are
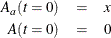
where x is the dosage.
The expected concentration of the substance in the body is computed by dividing the solution to the ODE, 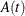 , by Cl, the clearance,
, by Cl, the clearance,

where i is the subject index.
Pinheiro and Bates (1995) consider the following first-order compartment model for these data, where 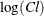 and
and 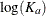 are modeled using random effects to account for the patient-to-patient variability:
are modeled using random effects to account for the patient-to-patient variability:
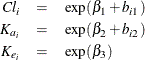
Here the 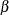 s denote fixed-effects parameters and the
s denote fixed-effects parameters and the 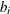 s denote random-effects parameters with an unknown covariance matrix.
s denote random-effects parameters with an unknown covariance matrix.
Although there is an analytical solution to this set of differential equations, this example illustrates the use of a general
ODE solver that does not require a closed-form solution. To use the ODE solver, you want to first define the set of differential
equations in PROC FCMP and store the objective function, called OneComp, in a user library:
proc fcmp outlib=sasuser.funcs.PK; subroutine OneComp(t,y[*],dy[*],ka,ke); outargs dy; dy[1] = -ka*y[1]; dy[2] = ka*y[1]-ke*y[2]; endsub; run;
The first argument of the OneComp subroutine is the time variable in the differential equation. The second argument is the with-respect-to variable, which
can be an array in case a multidimensional problem is required. The third argument is an array that stores the differential
equations. This dy argument must also be the OUTARGS variable in the subroutine. The subsequent variables are additional variables, depending
on the problem at hand.
In the OneComp subroutine, you can define 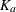 and
and 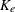 as functions of
as functions of 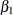 ,
, 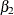 , and the random-effects parameter
, and the random-effects parameter 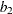 . The
. The dy[1] and dy[2] are the differential equation, with dy[1] storing 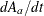 and
and dy[2] storing 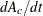 .
.
The following PROC MCMC statements use the CALL ODE
subroutine to solve the set of differential equations defined in OneComp and then use that solution in the construction of the likelihood function:
options cmplib=sasuser.funcs;
proc mcmc data=theoph nmc=10000 seed=27 outpost=theophO diag=none
nthreads=8;
ods select PostSumInt;
array b[2];
array muB[2] (0 0);
array cov[2,2];
array S[2,2] (1 0 0 1);
array init[2] dose 0;
array sol[2];
parms beta1 -3.22 beta2 0.47 beta3 -2.45 ;
parms cov {0.03 0 0 0.4};
parms s2y;
prior beta: ~ normal(0, sd=100);
prior cov ~ iwish(2, S);
prior s2y ~ igamma(shape=3, scale=2);
random b ~ mvn(muB, cov) subject=subject;
cl = exp(beta1 + b1);
ka = exp(beta2 + b2);
ke = exp(beta3);
v = cl/ke;
call ode('OneComp',sol,init,0,time,ka,ke);
mu = (sol[2]/v);
model conc ~ normal(mu,var=s2y);
run;
The INIT array stores the initial values of the two differential equations, with 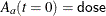 and
and 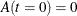 . The array is used as an input argument to the
. The array is used as an input argument to the CALL ODE subroutine.
The RANDOM
statement specifies two-dimensional random effects, b, with a multivariate normal prior. The first random effect, b1, enters the model through the clearance variable, cl. The second random effect, b2, is part of the differential equations. The CALL ODE
subroutine solves the OneComp set of differential equations and returns the solution to the SOL array. The second array element, SOL[2], is the solution to 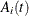 for every subject i at every time t.
for every subject i at every time t.
Posterior summary statistics are reported in Output 73.22.1.
Output 73.22.1: Posterior Summary Statistics
| Posterior Summaries and Intervals | |||||
|---|---|---|---|---|---|
| Parameter | N | Mean | Standard Deviation |
95% HPD Interval | |
| beta1 | 10000 | -3.2073 | 0.0836 | -3.3768 | -3.0429 |
| beta2 | 10000 | 0.4375 | 0.1860 | 0.1112 | 0.7671 |
| beta3 | 10000 | -2.4608 | 0.0547 | -2.5642 | -2.3576 |
| cov1 | 10000 | 0.1312 | 0.0637 | 0.0429 | 0.2507 |
| cov2 | 10000 | -0.00248 | 0.0919 | -0.1876 | 0.1793 |
| cov3 | 10000 | -0.00248 | 0.0919 | -0.1876 | 0.1793 |
| cov4 | 10000 | 0.6155 | 0.3175 | 0.1897 | 1.2190 |
| s2y | 10000 | 0.5246 | 0.0719 | 0.3954 | 0.6714 |
In this problem, the closed-form solution of the ODE is known:
![\[ C_{it} = \frac{D k_{e_ i} k_{a_ i}}{Cl_ i(k_{a_ i} - k_{e_ i})} [\exp (-k_{e_ i} t) - \exp (-k_{a_ i} t)] + e_{it} \]](images/statug_mcmc0859.png)
You can manually enter the equation in PROC MCMC and use the following program to fit the same model:
proc mcmc data=theoph nmc=10000 seed=22 outpost=theophC;
array b[2];
array mu[2] (0 0);
array cov[2,2];
array S[2,2] (1 0 0 1);
parms beta1 -3.22 beta2 0.47 beta3 -2.45 ;
parms cov {0.03 0 0 0.4};
parms s2y;
prior beta: ~ normal(0, sd=100);
prior cov ~ iwish(2, S);
prior s2y ~ igamma(shape=3, scale=2);
random b ~ mvn(mu, cov) subject=subject;
cl = exp(beta1 + b1);
ka = exp(beta2 + b2);
ke = exp(beta3);
pred = dose*ke*ka*(exp(-ke*time)-exp(-ka*time))/cl/(ka-ke);
model conc ~ normal(pred,var=s2y);
run;
Because this program makes it unnecessary to numerically solve the ODE at every observation and every iteration, it runs much faster than the program that uses the CALL ODE subroutine. But few pharmacokinetic models have known solutions that enable you to do that.