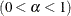The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
How PROC MCMC WorksBlocking of ParametersSampling MethodsTuning the Proposal DistributionDirect SamplingConjugate SamplingInitial Values of the Markov ChainsAssignments of ParametersStandard DistributionsUsage of Multivariate DistributionsSpecifying a New DistributionUsing Density Functions in the Programming StatementsTruncation and CensoringSome Useful SAS FunctionsMatrix Functions in PROC MCMCCreate Design MatrixModeling Joint LikelihoodAccess Lag and Lead VariablesCALL ODE and CALL QUAD SubroutinesRegenerating Diagnostics PlotsCaterpillar PlotAutocall Macros for PostprocessingGamma and Inverse-Gamma DistributionsPosterior Predictive DistributionHandling of Missing DataFunctions of Random-Effects ParametersFloating Point Errors and OverflowsHandling Error MessagesComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data
Simulating Samples From a Known DensityBox-Cox TransformationLogistic Regression Model with a Diffuse PriorLogistic Regression Model with Jeffreys’ PriorPoisson RegressionNonlinear Poisson Regression ModelsLogistic Regression Random-Effects ModelNonlinear Poisson Regression Multilevel Random-Effects ModelMultivariate Normal Random-Effects ModelMissing at Random AnalysisNonignorably Missing Data (MNAR) AnalysisChange Point ModelsExponential and Weibull Survival AnalysisTime Independent Cox ModelTime Dependent Cox ModelPiecewise Exponential Frailty ModelNormal Regression with Interval CensoringConstrained AnalysisImplement a New Sampling AlgorithmUsing a Transformation to Improve MixingGelman-Rubin DiagnosticsOne-Compartment Model with Pharmacokinetic Data - References
Autocall Macros for Postprocessing
Although PROC MCMC provides a number of convergence diagnostic tests and posterior summary statistics, PROC MCMC performs the calculations only if you specify the options in advance. If you wish to analyze the posterior draws of unmonitored parameters or functions of the parameters that are calculated in later DATA step calls, you can use the autocall macros in Table 73.46.
Table 73.46: Postprocessing Autocall Macros
|
Macro |
Description |
|
%ESS |
Effective sample sizes |
|
%GEWEKE* |
Geweke diagnostic |
|
%HEIDEL* |
Heidelberger-Welch diagnostic |
|
%MCSE |
Monte Carlo standard errors |
|
%RAFTERY |
Raftery diagnostic |
|
%POSTACF |
Autocorrelation |
|
%POSTCOR |
Correlation matrix |
|
%POSTCOV |
Covariance matrix |
|
%POSTINT |
Equal-tail and HPD intervals |
|
%POSTSUM |
Summary statistics |
|
%SUMINT |
Mean, standard deviation, and HPD interval |
|
*The %GEWEKE and %HEIDEL macros use a different optimization routine than that used in PROC MCMC. As a result, there might be numerical differences in some cases, especially when the sample size is small. |
|
Table 73.47 lists options that are shared by all postprocessing autocall macros. See Table 73.48 for macro-specific options.
Table 73.47: Shared Options
|
Option |
Description |
|---|---|
|
DATA=SAS-data-set |
Input data set that contains posterior samples |
|
VAR=variable-list |
Specifies the variables on which you want to carry out the calculation. |
|
PRINT=YES | NO |
Displays the results. The default is YES. |
|
OUT=SAS-data-set |
Specifies a name for the output SAS data set to contain the results. |
Suppose that the data set that contains posterior samples is called post and that the variables of interest are defined in the macro variable &PARMS. The following statements call the %ESS macro
and calculates the effective sample sizes for each variable:
%let parms = alpha beta u_1-u_17; %ESS(data=post, var=&parms);
By default, the ESS estimates are displayed. You can choose not to display the result and save the output to a data set with the following statement:
%ESS(data=post, var=&parms, print=NO, out=eout);
Some of the macros can take additional options, which are listed in Table 73.48.
Table 73.48: Macro-Specific Options
|
Macro |
Option |
Description |
|---|---|---|
|
%ESS |
AUTOCORLAG=numeric |
Specifies the maximum number of autocorrelation lags used in computing the ESS estimates. By default, AUTOCORLAG=MIN(500, NOBS/4), where NOBS is the sample size of the input data set. |
|
HIST=YES|NO |
Displays a histogram of all ESS estimates. The default is NO. |
|
|
%HEIDEL |
SALPHA=numeric |
Specifies the |
|
HALPHA=numeric |
Specifies the |
|
|
EPS=numeric |
Specifies a small positive number |
|
|
%GEWEKE |
FRAC1=numeric |
Specifies the earlier portion of the Markov chain used in the test. By default, FRAC1=0.1. |
|
FRAC2=numeric |
Specifies the latter portion of the Markov chain used in the test. By default, FRAC2=0.5. |
|
|
%MCSE |
AUTOCORLAG=numeric |
Specifies the maximum number of autocorrelation lags used in computing the Monte Carlo standard error estimates. By default, AUTOCORLAG=MIN(500, NOBS/4), where NOBS is the sample size of the input data set. |
|
%RAFTERY |
Q=numeric |
Specifies the order of the quantile of interest. By default, Q=0.025. |
|
R=numeric |
Specifies the margin of error for measuring the accuracy of estimation of the quantile. By default, R=0.005. |
|
|
S=numeric |
Specifies the probability of attaining the accuracy of the estimation of the quantile. By default, S=0.95. |
|
|
EPS=numeric |
Specifies the tolerance level for the stationary test. By default, EPS=0.001. |
|
|
%POSTACF |
LAGS=%str(numeric-list) |
Specifies autocorrelation lags calculated. The default values are 1, 5, 10, and 50. |
|
%POSTINT |
ALPHA=value |
Specifies the |
For example, the following statement calculates and displays autocorrelation at lags 1, 6, 11, 50, and 100. Note that the lags in the numeric-list need to be separated by commas “,”.
%PostACF(data=post, var=&parms, lags=%str(1 to 15 by 5, 50, 100));