The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC Works Blocking of Parameters Sampling Methods Tuning the Proposal Distribution Conjugate Sampling Initial Values of the Markov Chains Assignments of Parameters Standard Distributions Usage of Multivariate Distributions Specifying a New Distribution Using Density Functions in the Programming Statements Truncation and Censoring Some Useful SAS Functions Matrix Functions in PROC MCMC Create Design Matrix Modeling Joint Likelihood Regenerating Diagnostics Plots Caterpillar Plot Posterior Predictive Distribution Handling of Missing Data Floating Point Errors and Overflows Handling Error Messages Computational Resources Displayed Output ODS Table Names ODS Graphics
How PROC MCMC Works Blocking of Parameters Sampling Methods Tuning the Proposal Distribution Conjugate Sampling Initial Values of the Markov Chains Assignments of Parameters Standard Distributions Usage of Multivariate Distributions Specifying a New Distribution Using Density Functions in the Programming Statements Truncation and Censoring Some Useful SAS Functions Matrix Functions in PROC MCMC Create Design Matrix Modeling Joint Likelihood Regenerating Diagnostics Plots Caterpillar Plot Posterior Predictive Distribution Handling of Missing Data Floating Point Errors and Overflows Handling Error Messages Computational Resources Displayed Output ODS Table Names ODS Graphics -
Examples
 Simulating Samples From a Known Density Box-Cox Transformation Logistic Regression Model with a Diffuse Prior Logistic Regression Model with Jeffreys’ Prior Poisson Regression Nonlinear Poisson Regression Models Logistic Regression Random-Effects Model Nonlinear Poisson Regression Random-Effects Model Multivariate Normal Random-Effects Model Change Point Models Exponential and Weibull Survival Analysis Time Independent Cox Model Time Dependent Cox Model Piecewise Exponential Frailty Model Normal Regression with Interval Censoring Constrained Analysis Implement a New Sampling Algorithm Using a Transformation to Improve Mixing Gelman-Rubin Diagnostics
Simulating Samples From a Known Density Box-Cox Transformation Logistic Regression Model with a Diffuse Prior Logistic Regression Model with Jeffreys’ Prior Poisson Regression Nonlinear Poisson Regression Models Logistic Regression Random-Effects Model Nonlinear Poisson Regression Random-Effects Model Multivariate Normal Random-Effects Model Change Point Models Exponential and Weibull Survival Analysis Time Independent Cox Model Time Dependent Cox Model Piecewise Exponential Frailty Model Normal Regression with Interval Censoring Constrained Analysis Implement a New Sampling Algorithm Using a Transformation to Improve Mixing Gelman-Rubin Diagnostics - References
| Using Density Functions in the Programming Statements |
Density Functions in PROC MCMC
PROC MCMC has a number of internally defined log-density functions for univariate and multivariate distributions. These functions have the basic form of LPDFdist(x, parm-list), where dist is the name of the distribution (see Table 54.39 for univariate distributions and Table 54.40 for multivariate distributions). The argument x is the random variable, and parm-list is the list of parameters.
In addition, the univariate functions allow for optional boundary arguments, such as LPDFdist(x, parm-list, <lower>, <upper>), where lower and upper are optional but positional boundary arguments. With the exception of the Bernoulli and uniform distribution, you can specify limits on all univariate distributions.
To set a lower bound on the normal density:
lpdfnorm(x, 0, 1, -2);
To set just an upper bound, specify a missing value for the lower bound argument:
lpdfnorm(x, 0, 1, ., 2);
Leaving both limits out gives you the unbounded density. You can also specify both bounds:
lpdfnorm(x, 0, 1); lpdfnorm(x, 0, 1, -3, 4);
See Table 54.39 for the function names of univariate distributions and Table 54.40 for multivariate distributions.
Distribution Name |
Function Call |
|---|---|
Beta |
lpdfbeta(x, a, b, <lower>, <upper>); |
Binary |
lpdfbern(x, p); |
Binomial |
lpdfbin(x, n, p, <lower>, <upper>); |
Cauchy |
lpdfcau(x, loc, scale, <lower>, <upper>); |
|
lpdfchisq(x, df, <lower>, <upper>); |
Exponential |
lpdfechisq(x, df, <lower>, <upper>); |
Exponential gamma |
lpdfegamma(x, sp, scale, <lower>, <upper>); |
Exponential exponential |
lpdfeexpon(x, scale, <lower>, <upper>); |
Exponential inverse |
lpdfeichisq(x, df, <lower>, <upper>); |
Exponential inverse-gamma |
lpdfeigamma(x, sp, scale, <lower>, <upper>); |
Exponential scaled inverse |
lpdfesichisq(x, df, scale, <lower>, <upper>); |
Exponential |
lpdfexpon(x, scale, <lower>, <upper>); |
Gamma |
lpdfgamma(x, sp, scale, <lower>, <upper>); |
Geometric |
lpdfgeo(x, p, <lower>, <upper>); |
Inverse |
lpdfichisq(x, df, <lower>, <upper>); |
Inverse-gamma |
lpdfigamma(x, sp, scale, <lower>, <upper>); |
Laplace |
lpdfdexp(x, loc, scale, <lower>, <upper>); |
Logistic |
lpdflogis(x, loc, scale, <lower>, <upper>); |
Lognormal |
lpdflnorm(x, loc, sd, <lower>, <upper>); |
Negative binomial |
lpdfnegbin(x, n, p, <lower>, <upper>); |
Normal |
lpdfnorm(x, mu, sd, <lower>, <upper>); |
Pareto |
lpdfpareto(x, sp, scale, <lower>, <upper>); |
Poisson |
lpdfpoi(x, mean, <lower>, <upper>); |
Scaled inverse |
lpdfsichisq(x, df, scale, <lower>, <upper>); |
t |
lpdft(x, mu, sd, df, <lower>, <upper>); |
Uniform |
lpdfunif(x, a, b); |
Wald |
lpdfwald(x, mean, scale, <lower>, <upper>); |
Weibull |
lpdfwei(x, loc, sp, scale, <lower>, <upper>); |
In the multivariate log-density functions, arrays must be used in place for the random variable and parameters in the model.
Distribution Name |
Function Call |
|---|---|
Dirichlet |
lpdfdirch(x_array, alpha_array); |
Inverse Wishart |
lpdfiwish(x_array, df, S_array); |
Multivariate normal |
lpdfmvn(x_array, mu_array, cov_array); |
Multinomial |
lpdfmnom(x_array, p_array); |
Standard Distributions, the LOGPDF Functions, and the LPDFdist Functions
Standard distributions listed in the section Standard Distributions are names only, and they can be used only in the MODEL, PRIOR, and HYPERPRIOR statements to specify either a prior distribution or a conditional distribution of the data given parameters. They do not return any values, and you cannot use them in the programming statements.
The LOGPDF functions are DATA step functions that compute the logarithm of various probability density (mass) functions. For example, logpdf("beta", x, 2, 15) returns the log of a beta density with parameters a = 2 and b = 15, evaluated at  . All the LOGPDF functions are supported in PROC MCMC.
. All the LOGPDF functions are supported in PROC MCMC.
The LPDFdist functions are unique to PROC MCMC. They compute the logarithm of various probability density (mass) functions. The functions are the same as the LOGPDF functions when it comes to calculating the log density. For example, lpdfbeta(x, 2, 15) returns the same value as logpdf("beta", x, 2, 15). The LPDFdist functions cover a greater class of probability density functions, and the univariate distribution functions take the optional but positional boundary arguments. There are no corresponding LCDFdist or LSDFdist functions in PROC MCMC. To work with the cumulative probability function or the survival functions, you need to use the LOGCDF and the LOGSDF DATA step functions.
Multivariate Density Functions in the Data Step
The DATA step has functions that compute the logarithm of the density of some multivariate distributions. You can also use them in PROC MCMC. For a complete listing of multivariate functions, see SAS Language Reference: Dictionary.
Some commonly used multivariate functions are as follows:
LOGMPDFNORMAL, the logarithm of the multivariate normal
LOGMPDFWISHART, the logarithm of the Wishart
LOGMPDFIWISHART, the logarithm of the inverted-Wishart
LOGMPDFDIR1, the logarithm of the Dirichlet distribution of Type I
LOGMPDFDIR2, the logarithm of the Dirichlet distribution of Type II
LOGMPDFMULTINOM, the logarithm of the multinomial
Other multivariate density functions include: LOGMPDFT (t distribution), LOGMPDFGAMMA (gamma distribution), LOGMPDFBETA1 (beta of type I), and LOGMPDFBETA2 (beta of type II).
Density Function Definition
LOGMPDFNORMAL
Let  be an
be an  -dimensional random vector with mean vector
-dimensional random vector with mean vector 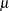 and covariance matrix
and covariance matrix 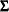 . The density is
. The density is
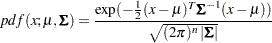 |
where 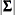 is the determinant of the covariance matrix
is the determinant of the covariance matrix 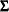 .
.
The function has syntax:
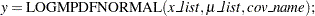 |
Warning: you must set up the 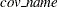 covariance matrix before using the LOGMPDFNORMAL function and free the memory after PROC MCMC exits. See the section Set Up the Covariance Matrices and Free Memory.
covariance matrix before using the LOGMPDFNORMAL function and free the memory after PROC MCMC exits. See the section Set Up the Covariance Matrices and Free Memory.
LOGMPDFWISHART and LOGMPDFIWISHART
The density function from the Wishart distribution is:
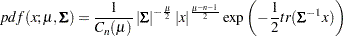 |
with 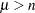 , and the trace of a square matrix
, and the trace of a square matrix  is given by:
is given by:
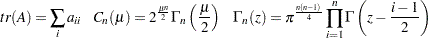 |
The density function from the inverse-Wishart distribution is:
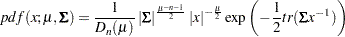 |
for 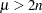 , and
, and
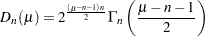 |
If  then
then 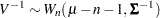
The functions have syntax:
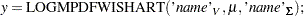 |
and for the inverted Wishart:
 |
The three arguments are the multivariate matrix 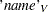 , the degrees of freedom
, the degrees of freedom 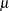 , and the covariance matrix
, and the covariance matrix 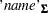 k
k
Warning: you must set up the 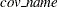 covariance matrix before using these functions and free the memory after PROC MCMC exits. See the section Set Up the Covariance Matrices and Free Memory.
covariance matrix before using these functions and free the memory after PROC MCMC exits. See the section Set Up the Covariance Matrices and Free Memory.
LOGMPDFDIR1 and LOGMPDFDIR2
The random variables 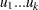 , with
, with 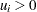 and
and 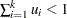 , are said to have a Dirichlet Type I distribution with parameters
, are said to have a Dirichlet Type I distribution with parameters 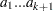 if their joint pdf is given by:
if their joint pdf is given by:
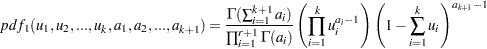 |
The variables are said to have a Dirichlet type II distribution with parameters 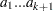 if their joint pdf is given by the following:
if their joint pdf is given by the following:
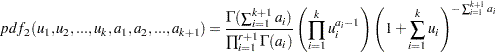 |
The functions have syntax:
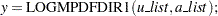 |
and
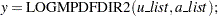 |
LOGMPDFMULTINOM
Let 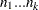 be random variables that denote the number of occurring of the events
be random variables that denote the number of occurring of the events 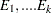 respectively occurring with probabilities
respectively occurring with probabilities  . Let
. Let 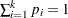 and let
and let 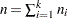 . Then the joint distribution of
. Then the joint distribution of 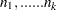 is the following:
is the following:
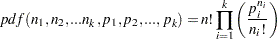 |
The function has syntax:
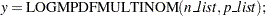 |
Set Up the Covariance Matrices and Free Memory
For distributions that require symmetric positive definite matrices, such as the LOGMPDFNORMAL, LOGMPDFWISHART and LOGMPDFIWISHART functions, you need to set up these matrices by using the following functions:
-
Use LOGMPDFSETSQ to set up a symmetric positive definite matrix from all its elements:
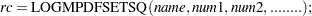
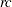 is set to
is set to 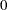 when the numeric arguments describe a symmetric positive definite matrix, otherwise it is set to a nonzero value.
when the numeric arguments describe a symmetric positive definite matrix, otherwise it is set to a nonzero value. -
Use LOGMPDFSET to set up a symmetric positive definite matrix from its lower triangular elements:
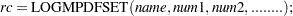
When the numeric arguments describe a symmetric positive definite matrix, the returned value
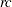 is set to
is set to 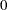 . Otherwise, a nonzero value for
. Otherwise, a nonzero value for 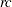 is returned.
is returned. -
Use LOGMPFFREE to free the workspace previously allocated with either LOGMPDFSET or LOGMPDFSETSQ:

When called without arguments, the LOGMPDFFREE frees all the symbols previously allocated by LOGMPDFSETSQ or LOGMPDFSET. Each freed symbol is reported back in the SAS log.
The parameters used in these functions are defined as follows:
is a string containing the name of the work space that stores the matrix by the numeric parameters
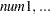 .
. are numeric arguments that represent the elements of a symmetric positive definite matrix.
You would set up this matrix under the DATA step by using the following syntax:
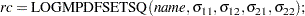 |
or the syntax:
 |
If the matrix is positive definite, the returned value 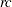 is zero.
is zero.