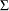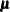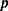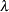The MCMC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 How PROC MCMC Works Blocking of Parameters Sampling Methods Tuning the Proposal Distribution Conjugate Sampling Initial Values of the Markov Chains Assignments of Parameters Standard Distributions Usage of Multivariate Distributions Specifying a New Distribution Using Density Functions in the Programming Statements Truncation and Censoring Some Useful SAS Functions Matrix Functions in PROC MCMC Create Design Matrix Modeling Joint Likelihood Regenerating Diagnostics Plots Caterpillar Plot Posterior Predictive Distribution Handling of Missing Data Floating Point Errors and Overflows Handling Error Messages Computational Resources Displayed Output ODS Table Names ODS Graphics
How PROC MCMC Works Blocking of Parameters Sampling Methods Tuning the Proposal Distribution Conjugate Sampling Initial Values of the Markov Chains Assignments of Parameters Standard Distributions Usage of Multivariate Distributions Specifying a New Distribution Using Density Functions in the Programming Statements Truncation and Censoring Some Useful SAS Functions Matrix Functions in PROC MCMC Create Design Matrix Modeling Joint Likelihood Regenerating Diagnostics Plots Caterpillar Plot Posterior Predictive Distribution Handling of Missing Data Floating Point Errors and Overflows Handling Error Messages Computational Resources Displayed Output ODS Table Names ODS Graphics -
Examples
 Simulating Samples From a Known Density Box-Cox Transformation Logistic Regression Model with a Diffuse Prior Logistic Regression Model with Jeffreys’ Prior Poisson Regression Nonlinear Poisson Regression Models Logistic Regression Random-Effects Model Nonlinear Poisson Regression Random-Effects Model Multivariate Normal Random-Effects Model Change Point Models Exponential and Weibull Survival Analysis Time Independent Cox Model Time Dependent Cox Model Piecewise Exponential Frailty Model Normal Regression with Interval Censoring Constrained Analysis Implement a New Sampling Algorithm Using a Transformation to Improve Mixing Gelman-Rubin Diagnostics
Simulating Samples From a Known Density Box-Cox Transformation Logistic Regression Model with a Diffuse Prior Logistic Regression Model with Jeffreys’ Prior Poisson Regression Nonlinear Poisson Regression Models Logistic Regression Random-Effects Model Nonlinear Poisson Regression Random-Effects Model Multivariate Normal Random-Effects Model Change Point Models Exponential and Weibull Survival Analysis Time Independent Cox Model Time Dependent Cox Model Piecewise Exponential Frailty Model Normal Regression with Interval Censoring Constrained Analysis Implement a New Sampling Algorithm Using a Transformation to Improve Mixing Gelman-Rubin Diagnostics - References
| Conjugate Sampling |
Conjugate prior is a family of prior distributions in which the prior and the posterior distributions are of the same family of distributions. For example, if you model an independently and identically distributed random variable 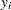 using a normal likelihood with known variance
using a normal likelihood with known variance 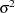 ,
,
 |
a normal prior on 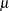
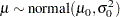 |
is a conjugate prior because the posterior distribution of 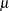 is also a normal distribution given
is also a normal distribution given 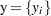 ,
, 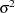 ,
,  , and
, and 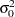 :
:
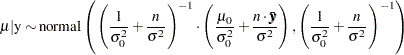 |
Conjugate sampling is efficient because it enables the Markov chain to obtain samples from the target distribution directly. When appropriate, PROC MCMC uses conjugate sampling methods to draw conditional posterior samples. Table 54.6 lists scenarios that lead to conjugate sampling in PROC MCMC.
Family |
Parameter |
Prior |
|---|---|---|
Normal with known |
Variance |
Inverse gamma family |
Normal with known |
Precision |
Gamma family |
Normal with known scale parameter ( |
Mean |
Normal |
Multivariate normal with known |
Mean |
Multivariate normal |
Multivariate normal with known |
Covariance |
Inverse Wishart |
Multinomial |
|
Dirichlet |
Binomial/binary |
|
Beta |
Poisson |
|
Gamma family |
In most cases, Family in Output 54.6 refers to the likelihood function. However, it does not necessarily have to be the case. The Family is a distribution that is conditional on the parameter of interest, and it can appear in any level of the hierarchical model, including on the random-effects level.
PROC MCMC can detect conjugacy only if the model parameter (not a function or a transformation of the model parameter) is used in the prior and Family distributions. For example, the following program leads to a conjugate sampler being used on the parameter mu:
parm mu; prior mu ~ n(0, sd=1000); model y ~ n(mu, var=s2);
However, if you modify the program slightly in the following way, although the conjugacy still holds in theory, PROC MCMC cannot detect conjugacy on mu because the parameter enters the normal likelihood function through the symbol w:
parm mu; prior mu ~ n(0, sd=1000); w = mu; model y ~ n(w, var=s2);
In this case, PROC MCMC resorts to the default sampling algorithm, which is a random walk Metropolis based on a normal kernel.
Similarly, the following statements also prevent PROC MCMC from detecting conjugacy on the parameter mu:
parm mu; prior mu ~ n(0, sd=1000); model y ~ n(mu + 2, var=s2);
When conjugacy is defected in a model, PROC MCMC performs a numerical optimization on the joint posterior distribution at the start of the MCMC simulation. To turn off this pre-optimization routine, use option PROPCOV=IND.
In a normal family, an often-used conjugate prior on the variance 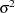 is
is
igamma(shape=0.001, scale=0.001)
An often-used conjugate prior on the precision  is
is
gamma(shape=0.001, iscale=0.001)
You want to exercise caution in using the igamma and gamma distributions as PROC MCMC suports both scale and iscale parametrizations in these distributions.
 , or
, or