The OPTGRAPH Procedure
- Overview
-
Getting Started

-
Syntax
 Functional SummaryPROC OPTGRAPH StatementBICONCOMP StatementCENTRALITY StatementCLIQUE StatementCOMMUNITY StatementCONCOMP StatementCORE StatementCYCLE StatementDATA_LINKS_VAR StatementDATA_MATRIX_VAR StatementDATA_NODES_VAR StatementEIGENVECTOR StatementLINEAR_ASSIGNMENT StatementMINCOSTFLOW StatementMINCUT StatementMINSPANTREE StatementPERFORMANCE StatementREACH StatementSHORTPATH StatementSUMMARY StatementTRANSITIVE_CLOSURE StatementTSP Statement
Functional SummaryPROC OPTGRAPH StatementBICONCOMP StatementCENTRALITY StatementCLIQUE StatementCOMMUNITY StatementCONCOMP StatementCORE StatementCYCLE StatementDATA_LINKS_VAR StatementDATA_MATRIX_VAR StatementDATA_NODES_VAR StatementEIGENVECTOR StatementLINEAR_ASSIGNMENT StatementMINCOSTFLOW StatementMINCUT StatementMINSPANTREE StatementPERFORMANCE StatementREACH StatementSHORTPATH StatementSUMMARY StatementTRANSITIVE_CLOSURE StatementTSP Statement -
Details
 Graph Input DataMatrix Input DataData Input OrderParallel ProcessingNumeric LimitationsSize LimitationsCommon Notation and AssumptionsBiconnected Components and Articulation PointsCentralityCliqueCommunityConnected ComponentsCore DecompositionCycleEigenvector ProblemLinear Assignment (Matching)Minimum-Cost Network FlowMinimum CutMinimum Spanning TreeReach (Ego) NetworkShortest PathSummaryTransitive ClosureTraveling Salesman ProblemMacro VariablesODS Table Names
Graph Input DataMatrix Input DataData Input OrderParallel ProcessingNumeric LimitationsSize LimitationsCommon Notation and AssumptionsBiconnected Components and Articulation PointsCentralityCliqueCommunityConnected ComponentsCore DecompositionCycleEigenvector ProblemLinear Assignment (Matching)Minimum-Cost Network FlowMinimum CutMinimum Spanning TreeReach (Ego) NetworkShortest PathSummaryTransitive ClosureTraveling Salesman ProblemMacro VariablesODS Table Names -
Examples
 Articulation Points in a Terrorist NetworkInfluence Centrality for Project Groups in a Research DepartmentBetweenness and Closeness Centrality for Computer Network TopologyBetweenness and Closeness Centrality for Project Groups in a Research DepartmentEigenvector Centrality for Word Sense DisambiguationCentrality Metrics for Project Groups in a Research DepartmentCommunity Detection on Zachary’s Karate Club DataRecursive Community Detection on Zachary’s Karate Club DataCycle Detection for Kidney Donor ExchangeLinear Assignment Problem for Minimizing Swim TimesLinear Assignment Problem, Sparse Format versus Dense FormatMinimum Spanning Tree for Computer Network TopologyTransitive Closure for Identification of Circular Dependencies in a Bug Tracking SystemReach Networks for Computation of Market Coverage of a Terrorist NetworkTraveling Salesman Tour through US Capital Cities
Articulation Points in a Terrorist NetworkInfluence Centrality for Project Groups in a Research DepartmentBetweenness and Closeness Centrality for Computer Network TopologyBetweenness and Closeness Centrality for Project Groups in a Research DepartmentEigenvector Centrality for Word Sense DisambiguationCentrality Metrics for Project Groups in a Research DepartmentCommunity Detection on Zachary’s Karate Club DataRecursive Community Detection on Zachary’s Karate Club DataCycle Detection for Kidney Donor ExchangeLinear Assignment Problem for Minimizing Swim TimesLinear Assignment Problem, Sparse Format versus Dense FormatMinimum Spanning Tree for Computer Network TopologyTransitive Closure for Identification of Circular Dependencies in a Bug Tracking SystemReach Networks for Computation of Market Coverage of a Terrorist NetworkTraveling Salesman Tour through US Capital Cities - References
Example 1.11 Linear Assignment Problem, Sparse Format versus Dense Format
This example looks at the problem of assigning swimmers to strokes based on their best times. However, in this case certain swimmers are not eligible to perform certain strokes. A missing (.) value in the data matrix identifies an ineligible assignment. For example:
data RelayTimesMatrix; input name $ sex $ back breast fly free; datalines; Sue F . 36.7 28.3 36.1 Karen F 34.6 . . 26.2 Jan F 31.3 . 27.1 . Andrea F 28.6 . 29.1 . Carol F 32.9 . 26.6 . ;
Recall that the linear assignment problem can also be interpreted as the minimum-weight matching in a bipartite graph. The eligible assignments define links between the rows (swimmers) and the columns (strokes), as in Figure 1.149.
Figure 1.149: Bipartite Graph for Linear Assignment Problem
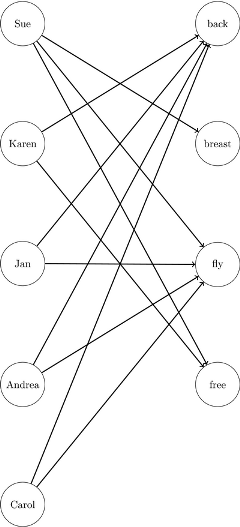
You can represent the same data in RelayTimesMatrix by using a links data set as follows:
data RelayTimesLinks; input name $ attr $ cost; datalines; Sue breast 36.7 Sue fly 28.3 Sue free 36.1 Karen back 34.6 Karen free 26.2 Jan back 31.3 Jan fly 27.1 Andrea back 28.6 Andrea fly 29.1 Carol back 32.9 Carol fly 26.6 ;
This graph must be bipartite (such that S and T are disjoint). If it is not, PROC OPTGRAPH returns an error.
Now, you can use either input format to solve the same problem, as follows:
proc optgraph
data_matrix = RelayTimesMatrix;
linear_assignment
out = LinearAssignMatrix
weight = (back--free)
id = (name sex);
run;
proc optgraph
graph_direction = directed
data_links = RelayTimesLinks;
data_links_var
from = name
to = attr
weight = cost;
linear_assignment
out = LinearAssignLinks;
run;
When you use the graph input format, the LINEAR_ASSIGNMENT options WEIGHT= and ID= are not used directly.
The data sets LinearAssignMatrix and LinearAssignLinks now contain the optimal assignments, as shown in Output 1.11.1 and Output 1.11.2.
Output 1.11.1: Optimal Assignments for Swim Times (Dense Input)
Output 1.11.2: Optimal Assignments for Swim Times (Sparse Input)
The optimal assignments are shown graphically in Figure 1.150.
Figure 1.150: Optimal Assignments for Swim Times
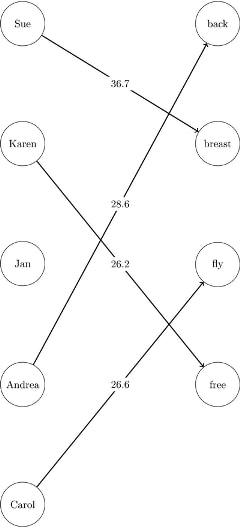
For large problems where a number of links are forbidden, the sparse format can be faster and can save a great deal of memory.
Consider an example that uses the format of the DATA_MATRIX= option with 15,000 columns (![]() ) and 4,000 rows (
) and 4,000 rows (![]() ). To store the dense matrix in memory, PROC OPTGRAPH needs to allocate approximately
). To store the dense matrix in memory, PROC OPTGRAPH needs to allocate approximately ![]() MB. If the data have mostly ineligible links, then the sparse (graph) format that uses the DATA_LINKS= option is much more
efficient with respect to memory. For example, if the data have only 5% of the eligible links (
MB. If the data have mostly ineligible links, then the sparse (graph) format that uses the DATA_LINKS= option is much more
efficient with respect to memory. For example, if the data have only 5% of the eligible links (![]() ), then the dense storage would still need 457 MB. The sparse storage for the same example needs approximately
), then the dense storage would still need 457 MB. The sparse storage for the same example needs approximately ![]() MB. If the problem is fully dense (all links are eligible), then the dense format that uses the DATA_MATRIX= option is the
most efficient.
MB. If the problem is fully dense (all links are eligible), then the dense format that uses the DATA_MATRIX= option is the
most efficient.