The OPTGRAPH Procedure
- Overview
-
Getting Started

-
Syntax
 Functional SummaryPROC OPTGRAPH StatementBICONCOMP StatementCENTRALITY StatementCLIQUE StatementCOMMUNITY StatementCONCOMP StatementCORE StatementCYCLE StatementDATA_LINKS_VAR StatementDATA_MATRIX_VAR StatementDATA_NODES_VAR StatementEIGENVECTOR StatementLINEAR_ASSIGNMENT StatementMINCOSTFLOW StatementMINCUT StatementMINSPANTREE StatementPERFORMANCE StatementREACH StatementSHORTPATH StatementSUMMARY StatementTRANSITIVE_CLOSURE StatementTSP Statement
Functional SummaryPROC OPTGRAPH StatementBICONCOMP StatementCENTRALITY StatementCLIQUE StatementCOMMUNITY StatementCONCOMP StatementCORE StatementCYCLE StatementDATA_LINKS_VAR StatementDATA_MATRIX_VAR StatementDATA_NODES_VAR StatementEIGENVECTOR StatementLINEAR_ASSIGNMENT StatementMINCOSTFLOW StatementMINCUT StatementMINSPANTREE StatementPERFORMANCE StatementREACH StatementSHORTPATH StatementSUMMARY StatementTRANSITIVE_CLOSURE StatementTSP Statement -
Details
 Graph Input DataMatrix Input DataData Input OrderParallel ProcessingNumeric LimitationsSize LimitationsCommon Notation and AssumptionsBiconnected Components and Articulation PointsCentralityCliqueCommunityConnected ComponentsCore DecompositionCycleEigenvector ProblemLinear Assignment (Matching)Minimum-Cost Network FlowMinimum CutMinimum Spanning TreeReach (Ego) NetworkShortest PathSummaryTransitive ClosureTraveling Salesman ProblemMacro VariablesODS Table Names
Graph Input DataMatrix Input DataData Input OrderParallel ProcessingNumeric LimitationsSize LimitationsCommon Notation and AssumptionsBiconnected Components and Articulation PointsCentralityCliqueCommunityConnected ComponentsCore DecompositionCycleEigenvector ProblemLinear Assignment (Matching)Minimum-Cost Network FlowMinimum CutMinimum Spanning TreeReach (Ego) NetworkShortest PathSummaryTransitive ClosureTraveling Salesman ProblemMacro VariablesODS Table Names -
Examples
 Articulation Points in a Terrorist NetworkInfluence Centrality for Project Groups in a Research DepartmentBetweenness and Closeness Centrality for Computer Network TopologyBetweenness and Closeness Centrality for Project Groups in a Research DepartmentEigenvector Centrality for Word Sense DisambiguationCentrality Metrics for Project Groups in a Research DepartmentCommunity Detection on Zachary’s Karate Club DataRecursive Community Detection on Zachary’s Karate Club DataCycle Detection for Kidney Donor ExchangeLinear Assignment Problem for Minimizing Swim TimesLinear Assignment Problem, Sparse Format versus Dense FormatMinimum Spanning Tree for Computer Network TopologyTransitive Closure for Identification of Circular Dependencies in a Bug Tracking SystemReach Networks for Computation of Market Coverage of a Terrorist NetworkTraveling Salesman Tour through US Capital Cities
Articulation Points in a Terrorist NetworkInfluence Centrality for Project Groups in a Research DepartmentBetweenness and Closeness Centrality for Computer Network TopologyBetweenness and Closeness Centrality for Project Groups in a Research DepartmentEigenvector Centrality for Word Sense DisambiguationCentrality Metrics for Project Groups in a Research DepartmentCommunity Detection on Zachary’s Karate Club DataRecursive Community Detection on Zachary’s Karate Club DataCycle Detection for Kidney Donor ExchangeLinear Assignment Problem for Minimizing Swim TimesLinear Assignment Problem, Sparse Format versus Dense FormatMinimum Spanning Tree for Computer Network TopologyTransitive Closure for Identification of Circular Dependencies in a Bug Tracking SystemReach Networks for Computation of Market Coverage of a Terrorist NetworkTraveling Salesman Tour through US Capital Cities - References
Example 1.9 Cycle Detection for Kidney Donor Exchange
This example looks at an application of cycle detection to help create a kidney donor exchange. Suppose someone needs a kidney transplant and a family member is willing to donate one. If the donor and recipient are incompatible (because of blood types, tissue mismatch, and so on), the transplant cannot happen. Now suppose two donor-recipient pairs A and B are in this situation, but donor A is compatible with recipient B and donor B is compatible with recipient A. Then two transplants can take place in a two-way swap, shown graphically in Figure 1.148. More generally, an n-way swap can be performed involving n donors and n recipients (Willingham 2009).
Figure 1.148: Kidney Donor Exchange Two-Way Swap
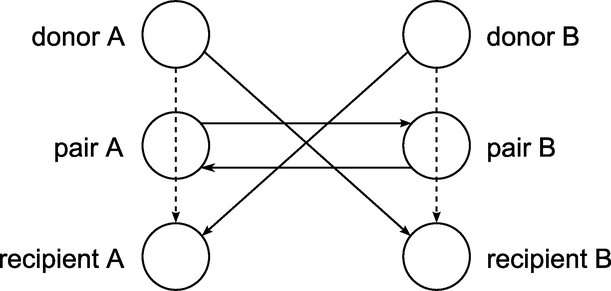
To model this problem, define a directed graph as follows. Each node is an incompatible donor-recipient pair. Link ![]() exists if the donor from node i is compatible with the recipient from node j. The link weight is a measure of the quality of the match. By introducing dummy links whose weight is 0, you can also include
altruistic donors who have no recipients or recipients who have no donors. The idea is to find a maximum-weight node-disjoint
union of directed cycles. You want the union to be node-disjoint so that no kidney is donated more than once, and you want
cycles so that the donor from node i gives up a kidney if and only if the recipient from node i receives a kidney.
exists if the donor from node i is compatible with the recipient from node j. The link weight is a measure of the quality of the match. By introducing dummy links whose weight is 0, you can also include
altruistic donors who have no recipients or recipients who have no donors. The idea is to find a maximum-weight node-disjoint
union of directed cycles. You want the union to be node-disjoint so that no kidney is donated more than once, and you want
cycles so that the donor from node i gives up a kidney if and only if the recipient from node i receives a kidney.
Without any other constraints, the problem could be solved as a linear assignment problem, as described in the section Linear Assignment (Matching). But doing so would allow arbitrarily long cycles in the solution. Because of practical considerations (such as travel) and to mitigate risk, each cycle must have no more than L links. The kidney exchange problem is to find a maximum-weight node-disjoint union of short directed cycles.
One way to solve this problem is to explicitly generate all cycles whose length is at most L and then solve a set packing problem. You can use PROC OPTGRAPH to generate the cycles and then PROC OPTMODEL (see SAS/OR User's Guide: Mathematical Programming) to read the PROC OPTGRAPH output, formulate the set packing problem, call the mixed integer linear programming solver, and output the optimal solution.
The following DATA step sets up the problem, first creating a random graph on n nodes with link probability p and Uniform(0,1) weight:
/* create random graph on n nodes with arc probability p
and uniform(0,1) weight */
%let n = 100;
%let p = 0.02;
data LinkSetIn;
do from = 0 to &n - 1;
do to = 0 to &n - 1;
if from eq to then continue;
else if ranuni(1) < &p then do;
weight = ranuni(2);
output;
end;
end;
end;
run;
The following statements use PROC OPTGRAPH to generate all cycles whose length is greater than or equal to 2 and less than or equal to 10:
/* generate all cycles with 2 <= length <= max_length */
%let max_length = 10;
proc optgraph
loglevel = moderate
graph_direction = directed
data_links = LinkSetIn;
cycle
minLength = 2
maxLength = &max_length
out = Cycles
mode = all_cycles;
run;
%put &_OPTGRAPH_;
%put &_OPTGRAPH_CYCLE_;
PROC OPTGRAPH finds 224 cycles of the appropriate length, as shown in Output 1.9.1.
Output 1.9.1: Cycles for Kidney Donor Exchange PROC OPTGRAPH Log
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: Running OPTGRAPH version 14.1. |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: The OPTGRAPH procedure is executing in single-machine mode. |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: Reading the links data set. |
| NOTE: There were 194 observations read from the data set WORK.LINKSETIN. |
| NOTE: Data input used 0.01 (cpu: 0.00) seconds. |
| NOTE: Building the input graph storage used 0.00 (cpu: 0.00) seconds. |
| NOTE: The input graph storage is using 0.3 MBs of memory. |
| NOTE: The number of nodes in the input graph is 97. |
| NOTE: The number of links in the input graph is 194. |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: Processing cycle detection. |
| NOTE: The graph has 224 cycles. |
| NOTE: Processing cycle detection used 2.31 (cpu: 2.26) seconds. |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: Creating cycle data set output. |
| NOTE: Data output used 0.00 (cpu: 0.00) seconds. |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: ------------------------------------------------------------------------------------------ |
| NOTE: The data set WORK.CYCLES has 2124 observations and 3 variables. |
| STATUS=OK CYCLE=OK |
| STATUS=OK NUM_CYCLES=224 CPU_TIME=2.26 REAL_TIME=2.31 |
From the resulting data set Cycles, use the following DATA step to convert the cycles into one observation per arc:
/* convert Cycles into one observation per arc */ data Cycles0(keep=c i j); set Cycles; retain last; c = cycle; i = last; j = node; last = j; if order ne 1 then output; run;
Given the set of cycles, you can now formulate a mixed integer linear program (MILP) to maximize the total cycle weight. Let
C define the set of cycles of appropriate length, ![]() define the set of nodes in cycle c,
define the set of nodes in cycle c, ![]() define the set of links in cycle c, and
define the set of links in cycle c, and ![]() denote the link weight for link
denote the link weight for link ![]() . Define a binary decision variable
. Define a binary decision variable ![]() . Set
. Set ![]() to 1 if cycle c is used in the solution; otherwise, set it to 0. Then, the following MILP defines the problem that you want to solve to maximize
the quality of the kidney exchange:
to 1 if cycle c is used in the solution; otherwise, set it to 0. Then, the following MILP defines the problem that you want to solve to maximize
the quality of the kidney exchange:
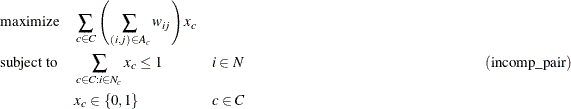
The constraint (incomp_pair) ensures that each node (incompatible pair) in the graph is intersected at most once. That is, a donor can donate a kidney only once. You can use PROC OPTMODEL to solve this mixed integer linear programming problem as follows:
/* solve set packing problem to find maximum weight node-disjoint union
of short directed cycles */
proc optmodel;
/* declare index sets and parameters, and read data */
set <num,num> ARCS;
num weight {ARCS};
read data LinkSetIn into ARCS=[from to] weight;
set <num,num,num> TRIPLES;
read data Cycles0 into TRIPLES=[c i j];
set CYCLES = setof {<c,i,j> in TRIPLES} c;
set ARCS_c {c in CYCLES} = setof {<(c),i,j> in TRIPLES} <i,j>;
set NODES_c {c in CYCLES} = union {<i,j> in ARCS_c[c]} {i,j};
set NODES = union {c in CYCLES} NODES_c[c];
num cycle_weight {c in CYCLES} = sum {<i,j> in ARCS_c[c]} weight[i,j];
/* UseCycle[c] = 1 if cycle c is used, 0 otherwise */
var UseCycle {CYCLES} binary;
/* declare objective */
max TotalWeight
= sum {c in CYCLES} cycle_weight[c] * UseCycle[c];
/* each node appears in at most one cycle */
con node_packing {i in NODES}:
sum {c in CYCLES: i in NODES_c[c]} UseCycle[c] <= 1;
/* call solver */
solve with milp;
/* output optimal solution */
create data Solution from
[c]={c in CYCLES: UseCycle[c].sol > 0.5} cycle_weight;
quit;
%put &_OROPTMODEL_;
PROC OPTMODEL solves the problem by using the mixed integer linear programming solver. As shown in Output 1.9.2, it was able to find a total weight (quality level) of 26.02.
Output 1.9.2: Cycles for Kidney Donor Exchange PROC OPTMODEL Log
| NOTE: There were 194 observations read from the data set WORK.LINKSETIN. |
| NOTE: There were 1900 observations read from the data set WORK.CYCLES0. |
| NOTE: Problem generation will use 4 threads. |
| NOTE: The problem has 224 variables (0 free, 0 fixed). |
| NOTE: The problem has 224 binary and 0 integer variables. |
| NOTE: The problem has 63 linear constraints (63 LE, 0 EQ, 0 GE, 0 range). |
| NOTE: The problem has 1900 linear constraint coefficients. |
| NOTE: The problem has 0 nonlinear constraints (0 LE, 0 EQ, 0 GE, 0 range). |
| NOTE: The MILP presolver value AUTOMATIC is applied. |
| NOTE: The MILP presolver removed 46 variables and 35 constraints. |
| NOTE: The MILP presolver removed 901 constraint coefficients. |
| NOTE: The MILP presolver modified 0 constraint coefficients. |
| NOTE: The presolved problem has 178 variables, 28 constraints, and 999 constraint coefficients. |
| NOTE: The MILP solver is called. |
| NOTE: The parallel Branch and Cut algorithm is used. |
| NOTE: The Branch and Cut algorithm is using up to 4 threads. |
| Node Active Sols BestInteger BestBound Gap Time |
| 0 1 3 22.7780692 868.9019355 97.38% 0 |
| 0 1 3 22.7780692 26.7803921 14.94% 0 |
| 0 1 4 23.2158345 26.1205372 11.12% 0 |
| 0 1 5 26.0202879 26.0202879 0.00% 0 |
| 0 0 5 26.0202879 26.0202879 0.00% 0 |
| NOTE: The MILP solver added 31 cuts with 2815 cut coefficients at the root. |
| NOTE: Objective = 26.020287887. |
| NOTE: The data set WORK.SOLUTION has 6 observations and 2 variables. |
| STATUS=OK ALGORITHM=BAC SOLUTION_STATUS=OPTIMAL OBJECTIVE=26.020287887 RELATIVE_GAP=0 |
| ABSOLUTE_GAP=0 PRIMAL_INFEASIBILITY=1.110223E-15 BOUND_INFEASIBILITY=0 |
| INTEGER_INFEASIBILITY=4E-6 BEST_BOUND=26.020287887 NODES=1 ITERATIONS=117 PRESOLVE_TIME=0.01 |
| SOLUTION_TIME=0.04 |
The data set Solution, shown in Output 1.9.3, now contains the cycles that define the best exchange and their associated weight (quality).
Output 1.9.3: Maximum Quality Solution for Kidney Donor Exchange