The OPTGRAPH Procedure
- Overview
-
Getting Started

-
Syntax
 Functional SummaryPROC OPTGRAPH StatementBICONCOMP StatementCENTRALITY StatementCLIQUE StatementCOMMUNITY StatementCONCOMP StatementCORE StatementCYCLE StatementDATA_LINKS_VAR StatementDATA_MATRIX_VAR StatementDATA_NODES_VAR StatementEIGENVECTOR StatementLINEAR_ASSIGNMENT StatementMINCOSTFLOW StatementMINCUT StatementMINSPANTREE StatementPERFORMANCE StatementREACH StatementSHORTPATH StatementSUMMARY StatementTRANSITIVE_CLOSURE StatementTSP Statement
Functional SummaryPROC OPTGRAPH StatementBICONCOMP StatementCENTRALITY StatementCLIQUE StatementCOMMUNITY StatementCONCOMP StatementCORE StatementCYCLE StatementDATA_LINKS_VAR StatementDATA_MATRIX_VAR StatementDATA_NODES_VAR StatementEIGENVECTOR StatementLINEAR_ASSIGNMENT StatementMINCOSTFLOW StatementMINCUT StatementMINSPANTREE StatementPERFORMANCE StatementREACH StatementSHORTPATH StatementSUMMARY StatementTRANSITIVE_CLOSURE StatementTSP Statement -
Details
 Graph Input DataMatrix Input DataData Input OrderParallel ProcessingNumeric LimitationsSize LimitationsCommon Notation and AssumptionsBiconnected Components and Articulation PointsCentralityCliqueCommunityConnected ComponentsCore DecompositionCycleEigenvector ProblemLinear Assignment (Matching)Minimum-Cost Network FlowMinimum CutMinimum Spanning TreeReach (Ego) NetworkShortest PathSummaryTransitive ClosureTraveling Salesman ProblemMacro VariablesODS Table Names
Graph Input DataMatrix Input DataData Input OrderParallel ProcessingNumeric LimitationsSize LimitationsCommon Notation and AssumptionsBiconnected Components and Articulation PointsCentralityCliqueCommunityConnected ComponentsCore DecompositionCycleEigenvector ProblemLinear Assignment (Matching)Minimum-Cost Network FlowMinimum CutMinimum Spanning TreeReach (Ego) NetworkShortest PathSummaryTransitive ClosureTraveling Salesman ProblemMacro VariablesODS Table Names -
Examples
 Articulation Points in a Terrorist NetworkInfluence Centrality for Project Groups in a Research DepartmentBetweenness and Closeness Centrality for Computer Network TopologyBetweenness and Closeness Centrality for Project Groups in a Research DepartmentEigenvector Centrality for Word Sense DisambiguationCentrality Metrics for Project Groups in a Research DepartmentCommunity Detection on Zachary’s Karate Club DataRecursive Community Detection on Zachary’s Karate Club DataCycle Detection for Kidney Donor ExchangeLinear Assignment Problem for Minimizing Swim TimesLinear Assignment Problem, Sparse Format versus Dense FormatMinimum Spanning Tree for Computer Network TopologyTransitive Closure for Identification of Circular Dependencies in a Bug Tracking SystemReach Networks for Computation of Market Coverage of a Terrorist NetworkTraveling Salesman Tour through US Capital Cities
Articulation Points in a Terrorist NetworkInfluence Centrality for Project Groups in a Research DepartmentBetweenness and Closeness Centrality for Computer Network TopologyBetweenness and Closeness Centrality for Project Groups in a Research DepartmentEigenvector Centrality for Word Sense DisambiguationCentrality Metrics for Project Groups in a Research DepartmentCommunity Detection on Zachary’s Karate Club DataRecursive Community Detection on Zachary’s Karate Club DataCycle Detection for Kidney Donor ExchangeLinear Assignment Problem for Minimizing Swim TimesLinear Assignment Problem, Sparse Format versus Dense FormatMinimum Spanning Tree for Computer Network TopologyTransitive Closure for Identification of Circular Dependencies in a Bug Tracking SystemReach Networks for Computation of Market Coverage of a Terrorist NetworkTraveling Salesman Tour through US Capital Cities - References
Core Decomposition
An alternative to community detection for detecting cohesive subgroups is a method for extracting k-cores, known as core decomposition. Although this method is generally not as powerful as community detection for extracting a detailed community structure,
it can give a coarse approximation of cohesive structure at a very low computational cost. Let ![]() define a graph with nodes N and links A, and let
define a graph with nodes N and links A, and let ![]() be an induced subgraph on nodes S. The subgraph
be an induced subgraph on nodes S. The subgraph ![]() is a k-core if and only if for every node
is a k-core if and only if for every node ![]() , the degree of v is greater than or equal to k and
, the degree of v is greater than or equal to k and ![]() is the maximum subgraph with this property. By definition, the cores are nested. That is, if
is the maximum subgraph with this property. By definition, the cores are nested. That is, if ![]() is a k-core of size k, then
is a k-core of size k, then ![]() is contained in
is contained in ![]() .
.
In PROC OPTGRAPH, you can invoke core decomposition by using the CORE statement. The options for this statement are described in the section CORE Statement.
The results for the core decomposition algorithm are given in the output node data set that is specified in the OUT_NODES=
option in the PROC OPTGRAPH statement. For each node in the node data set, the variable core_out identifies its core number, the highest-order core that contains this node. The core identifiers are numbered sequentially starting from 0.
The core decomposition algorithm reports status information in a macro variable called _OPTGRAPH_CORE_. See the section Macro Variable _OPTGRAPH_CORE_ for more information about this macro variable.
The algorithm used for core decomposition is based on the work presented in Batagelj and Zaversnik 2003. This algorithm runs in time ![]() and therefore should scale to very large graphs.
and therefore should scale to very large graphs.
Core Decomposition of a Simple Undirected Graph
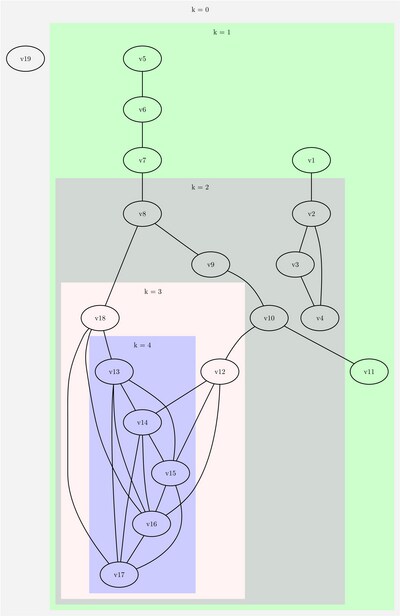
This section illustrates the use of the core decomposition algorithm on the simple undirected graph G that is shown in Figure 1.63.
Figure 1.63: Simple Undirected Graph
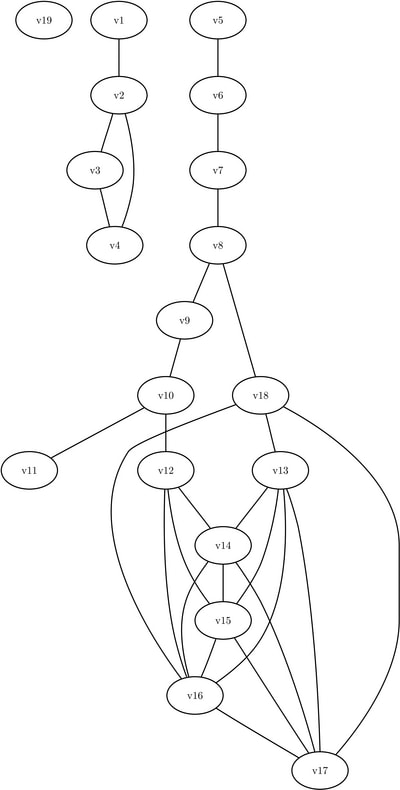
The undirected graph G can be represented using the following nodes data set NodeSetIn and links data set LinkSetIn:
data NodeSetIn; input node $ @@; datalines; v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 ; data LinkSetIn; input from $ to $ @@; datalines; v1 v2 v5 v6 v6 v7 v7 v8 v10 v11 v2 v3 v3 v4 v2 v4 v8 v9 v9 v10 v8 v18 v10 v12 v13 v14 v13 v15 v13 v16 v13 v17 v14 v15 v14 v16 v14 v17 v15 v16 v15 v17 v16 v17 v18 v13 v18 v17 v18 v16 v12 v14 v12 v15 v12 v16 ;
The following statements calculate the core decomposition and output the results in the data set NodeSetOut:
proc optgraph data_nodes = NodeSetIn data_links = LinkSetIn out_nodes = NodeSetOut; core; run;
The node data set NodeSetOut contains the core number (variable core_out) for each node and is shown in Figure 1.64.
Figure 1.64: Core Decomposition of a Simple Undirected Graph
Figure 1.65 shows the graph layered by its core number.
Figure 1.65: Core Decomposition