The MI Procedure
- Overview
- Getting Started
-
Syntax

-
Details
 Descriptive StatisticsEM Algorithm for Data with Missing ValuesStatistical Assumptions for Multiple ImputationMissing Data PatternsImputation MethodsMonotone Methods for Data Sets with Monotone Missing PatternsMonotone and FCS Regression MethodsMonotone and FCS Predictive Mean Matching MethodsMonotone and FCS Discriminant Function MethodsMonotone and FCS Logistic Regression MethodsMonotone Propensity Score MethodFCS Methods for Data Sets with Arbitrary Missing PatternsChecking Convergence in FCS MethodsMCMC Method for Arbitrary Missing Multivariate Normal DataProducing Monotone Missingness with the MCMC MethodMCMC Method SpecificationsChecking Convergence in MCMCInput Data SetsOutput Data SetsCombining Inferences from Multiply Imputed Data SetsMultiple Imputation EfficiencyNumber of ImputationsImputer’s Model Versus Analyst’s ModelParameter Simulation versus Multiple ImputationSensitivity Analysis for the MAR AssumptionMultiple Imputation with Pattern-Mixture ModelsSpecifying Sets of Observations for Imputation in Pattern-Mixture ModelsAdjusting Imputed Values in Pattern-Mixture ModelsSummary of Issues in Multiple ImputationPlot Options Superseded by ODS GraphicsODS Table NamesODS Graphics
Descriptive StatisticsEM Algorithm for Data with Missing ValuesStatistical Assumptions for Multiple ImputationMissing Data PatternsImputation MethodsMonotone Methods for Data Sets with Monotone Missing PatternsMonotone and FCS Regression MethodsMonotone and FCS Predictive Mean Matching MethodsMonotone and FCS Discriminant Function MethodsMonotone and FCS Logistic Regression MethodsMonotone Propensity Score MethodFCS Methods for Data Sets with Arbitrary Missing PatternsChecking Convergence in FCS MethodsMCMC Method for Arbitrary Missing Multivariate Normal DataProducing Monotone Missingness with the MCMC MethodMCMC Method SpecificationsChecking Convergence in MCMCInput Data SetsOutput Data SetsCombining Inferences from Multiply Imputed Data SetsMultiple Imputation EfficiencyNumber of ImputationsImputer’s Model Versus Analyst’s ModelParameter Simulation versus Multiple ImputationSensitivity Analysis for the MAR AssumptionMultiple Imputation with Pattern-Mixture ModelsSpecifying Sets of Observations for Imputation in Pattern-Mixture ModelsAdjusting Imputed Values in Pattern-Mixture ModelsSummary of Issues in Multiple ImputationPlot Options Superseded by ODS GraphicsODS Table NamesODS Graphics -
Examples
 EM Algorithm for MLEMonotone Propensity Score MethodMonotone Regression MethodMonotone Logistic Regression Method for CLASS VariablesMonotone Discriminant Function Method for CLASS VariablesFCS Methods for Continuous VariablesFCS Method for CLASS VariablesFCS Method with Trace PlotMCMC MethodProducing Monotone Missingness with MCMCChecking Convergence in MCMCSaving and Using Parameters for MCMCTransforming to NormalityMultistage ImputationCreating Control-Based Pattern Imputation in Sensitivity AnalysisAdjusting Imputed Continuous Values in Sensitivity AnalysisAdjusting Imputed Classification Levels in Sensitivity AnalysisAdjusting Imputed Values with Parameters in a Data Set
EM Algorithm for MLEMonotone Propensity Score MethodMonotone Regression MethodMonotone Logistic Regression Method for CLASS VariablesMonotone Discriminant Function Method for CLASS VariablesFCS Methods for Continuous VariablesFCS Method for CLASS VariablesFCS Method with Trace PlotMCMC MethodProducing Monotone Missingness with MCMCChecking Convergence in MCMCSaving and Using Parameters for MCMCTransforming to NormalityMultistage ImputationCreating Control-Based Pattern Imputation in Sensitivity AnalysisAdjusting Imputed Continuous Values in Sensitivity AnalysisAdjusting Imputed Classification Levels in Sensitivity AnalysisAdjusting Imputed Values with Parameters in a Data Set - References
EM Algorithm for Data with Missing Values
The EM algorithm (Dempster, Laird, and Rubin 1977) is a technique that finds maximum likelihood estimates in parametric models for incomplete data. For a detailed description and applications of the EM algorithm, see the books by Little and Rubin (2002); Schafer (1997); McLachlan and Krishnan (1997).
The EM algorithm is an iterative procedure that finds the MLE of the parameter vector by repeating the following steps:
1. The expectation E-step Given a set of parameter estimates, such as a mean vector and covariance matrix for a multivariate normal distribution, the E-step calculates the conditional expectation of the complete-data log likelihood given the observed data and the parameter estimates.
2. The maximization M-step Given a complete-data log likelihood, the M-step finds the parameter estimates to maximize the complete-data log likelihood from the E-step.
The two steps are iterated until the iterations converge.
In the EM process, the observed-data log likelihood is nondecreasing at each iteration. For multivariate normal data, suppose there are G groups with distinct missing patterns. Then the observed-data log likelihood being maximized can be expressed as
![\[ \mr{log} \, L(\theta | Y_{\mathit{obs}}) = \sum _{g=1}^{G} {\mr{log} \, L_{g}(\btheta | Y_{\mathit{obs}})} \]](images/statug_mi0069.png)
where 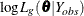 is the observed-data log likelihood from the gth group, and
is the observed-data log likelihood from the gth group, and
![\[ \mr{log} \, L_{g}(\btheta | Y_{\mathit{obs}}) = - {\frac{n_{g}}{2} } \, \mr{log} \, | {\Sigma }_{g} | - \, {\frac{1}{2} } \, \sum _{ig}{ (\mb{y}_{ig} - \bmu _ g)’ {{\bSigma }_{g}}^{-1} (\mb{y}_{ig} - \bmu _ g) } \]](images/statug_mi0071.png)
where 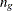 is the number of observations in the gth group, the summation is over observations in the gth group,
is the number of observations in the gth group, the summation is over observations in the gth group, 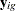 is a vector of observed values corresponding to observed variables,
is a vector of observed values corresponding to observed variables, 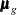 is the corresponding mean vector, and
is the corresponding mean vector, and 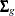 is the associated covariance matrix.
is the associated covariance matrix.
A sample covariance matrix is computed at each step of the EM algorithm. If the covariance matrix is singular, the linearly dependent variables for the observed data are excluded from the likelihood function. That is, for each observation with linear dependency among its observed variables, the dependent variables are excluded from the likelihood function. Note that this can result in an unexpected change in the likelihood between iterations prior to the final convergence.
See Schafer (1997, pp. 163–181) for a detailed description of the EM algorithm for multivariate normal data.
By default, PROC MI uses the means and standard deviations from available cases as the initial estimates for the EM algorithm. The correlations are set to zero. These estimates provide a good starting value with positive definite covariance matrix. For a discussion of suggested starting values for the algorithm, see Schafer (1997, p. 169).
You can specify the convergence criterion with the CONVERGE= option in the EM statement. The iterations are considered to have converged when the maximum change in the parameter estimates between iteration steps is less than the value specified. You can also specify the maximum number of iterations used in the EM algorithm with the MAXITER= option.
The MI procedure displays tables of the initial parameter estimates used to begin the EM process and the MLE parameter estimates
derived from EM. You can also display the EM iteration history with the ITPRINT option. PROC MI lists the iteration number,
the likelihood –2 log L, and the parameter values 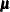 at each iteration. You can also save the MLE derived from the EM algorithm in a SAS data set by specifying the OUTEM= option.
at each iteration. You can also save the MLE derived from the EM algorithm in a SAS data set by specifying the OUTEM= option.