The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement
PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement -
Details
 Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsTweedie Distribution For Generalized Linear ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionResponse Level OrderingMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics
Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsTweedie Distribution For Generalized Linear ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionResponse Level OrderingMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics -
Examples
 Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson RegressionTweedie Regression
Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson RegressionTweedie Regression - References
Residuals
The GENMOD procedure computes three kinds of residuals. Residuals are available for all generalized linear models except multinomial models for ordinal response data, for which residuals are not available. Raw residuals and Pearson residuals are available for models fit with generalized estimating equations (GEEs).
The raw residual is defined as
![\[ r_ i = y_ i - \mu _ i \]](images/statug_genmod0292.png)
where 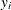 is the ith response and
is the ith response and 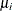 is the corresponding predicted mean. You can request raw residuals in an output data set with the keyword RESRAW
in the OUTPUT statement.
is the corresponding predicted mean. You can request raw residuals in an output data set with the keyword RESRAW
in the OUTPUT statement.
The Pearson residual is the square root of the ith contribution to the Pearson’s chi-square:
![\[ r_{Pi} = (y_ i - \mu _ i) \sqrt { \frac{w_ i}{V(\mu _ i)} } \]](images/statug_genmod0293.png)
You can request Pearson residuals in an output data set with the keyword RESCHI in the OUTPUT statement.
Finally, the deviance residual is defined as the square root of the contribution of the ith observation to the deviance, with the sign of the raw residual:
![\[ r_{Di} = \sqrt {d_ i}(\mr{sign}(y_ i - \mu _ i)) \]](images/statug_genmod0294.png)
You can request deviance residuals in an output data set with the keyword RESDEV in the OUTPUT statement.
The adjusted Pearson, deviance, and likelihood residuals are defined by Agresti (2002); Williams (1987); Davison and Snell (1991). These residuals are useful for outlier detection and for assessing the influence of single observations on the fitted model.
For the generalized linear model, the variance of the ith individual observation is given by
![\[ v_ i = \frac{\phi V(\mu _ i)}{w_ i} \]](images/statug_genmod0295.png)
where  is the dispersion parameter,
is the dispersion parameter, 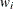 is a user-specified prior weight (if not specified,
is a user-specified prior weight (if not specified, 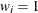 ),
), 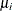 is the mean, and
is the mean, and 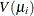 is the variance function. Let
is the variance function. Let
![\[ w_{ei} = v_ i^{-1}(g^{\prime }(\mu _ i))^{-2} \]](images/statug_genmod0297.png)
for the ith observation, where 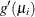 is the derivative of the link function, evaluated at
is the derivative of the link function, evaluated at 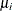 . Let
. Let 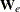 be the diagonal matrix with
be the diagonal matrix with 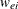 denoting the ith diagonal element. The weight matrix
denoting the ith diagonal element. The weight matrix 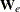 is used in computing the expected information matrix.
is used in computing the expected information matrix.
Define 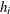 as the ith diagonal element of the matrix
as the ith diagonal element of the matrix
![\[ \mb{W}_ e^\frac {1}{2} \mb{X} (\mb{X}^{\prime } \mb{W}_ e \mb{X})^{-1} \mb{X}^{\prime } \mb{W}_ e^\frac {1}{2} \]](images/statug_genmod0300.png)
The Pearson residuals, standardized to have unit asymptotic variance, are given by
![\[ r_{Pi} = \frac{y_ i - \mu _ i}{\sqrt {v_ i(1 - h_ i)}} \]](images/statug_genmod0301.png)
You can request standardized Pearson residuals in an output data set with the keyword STDRESCHI in the OUTPUT statement. The deviance residuals, standardized to have unit asymptotic variance, are given by
![\[ r_{Di} = \frac{\mr{sign}(y_ i - \mu _ i) \sqrt {d_ i}}{\sqrt {\phi (1 - h_ i)}} \]](images/statug_genmod0302.png)
where 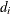 is the contribution to the total deviance from observation i, and
is the contribution to the total deviance from observation i, and 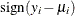 is 1 if
is 1 if 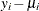 is positive and –1 if
is positive and –1 if 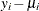 is negative. You can request standardized deviance residuals in an output data set with the keyword STDRESDEV
in the OUTPUT statement. The likelihood
residuals are defined by
is negative. You can request standardized deviance residuals in an output data set with the keyword STDRESDEV
in the OUTPUT statement. The likelihood
residuals are defined by
![\[ r_{Gi} = \mr{sign}(y_ i - \mu _ i) \sqrt {(1 - h_ i)r_{Di}^2 + h_ i r_{Pi}^2} \]](images/statug_genmod0306.png)
You can request likelihood residuals in an output data set with the keyword RESLIK in the OUTPUT statement.