The CALIS Procedure
-
Overview

-
Getting Started

-
Syntax
 Classes of Statements in PROC CALIS Single-Group Analysis Syntax Multiple-Group Multiple-Model Analysis Syntax PROC CALIS Statement BOUNDS Statement BY Statement COSAN Statement COV Statement DETERM Statement EFFPART Statement FACTOR Statement FITINDEX Statement FREQ Statement GROUP Statement LINCON Statement LINEQS Statement LISMOD Statement LMTESTS Statement MATRIX Statement MEAN Statement MODEL Statement MSTRUCT Statement NLINCON Statement NLOPTIONS Statement OUTFILES Statement PARAMETERS Statement PARTIAL Statement PATH Statement PCOV Statement PVAR Statement RAM Statement REFMODEL Statement RENAMEPARM Statement SAS Programming Statements SIMTESTS Statement STD Statement STRUCTEQ Statement TESTFUNC Statement VAR Statement VARIANCE Statement VARNAMES Statement WEIGHT Statement
Classes of Statements in PROC CALIS Single-Group Analysis Syntax Multiple-Group Multiple-Model Analysis Syntax PROC CALIS Statement BOUNDS Statement BY Statement COSAN Statement COV Statement DETERM Statement EFFPART Statement FACTOR Statement FITINDEX Statement FREQ Statement GROUP Statement LINCON Statement LINEQS Statement LISMOD Statement LMTESTS Statement MATRIX Statement MEAN Statement MODEL Statement MSTRUCT Statement NLINCON Statement NLOPTIONS Statement OUTFILES Statement PARAMETERS Statement PARTIAL Statement PATH Statement PCOV Statement PVAR Statement RAM Statement REFMODEL Statement RENAMEPARM Statement SAS Programming Statements SIMTESTS Statement STD Statement STRUCTEQ Statement TESTFUNC Statement VAR Statement VARIANCE Statement VARNAMES Statement WEIGHT Statement -
Details
 Input Data Sets Output Data Sets The COSAN Model The FACTOR Model The LINEQS Model The LISMOD Model and Submodels The MSTRUCT Model The PATH Model The RAM Model Naming Variables and Parameters Setting Constraints on Parameters Automatic Variable Selection Estimation Criteria Relationships among Estimation Criteria Gradient, Hessian, Information Matrix, and Approximate Standard Errors Counting the Degrees of Freedom Assessment of Fit Total, Direct, and Indirect Effects Standardized Solutions Modification Indices Missing Values and the Analysis of Missing Patterns Measures of Multivariate Kurtosis Initial Estimates Use of Optimization Techniques Computational Problems Displayed Output ODS Table Names ODS Graphics
Input Data Sets Output Data Sets The COSAN Model The FACTOR Model The LINEQS Model The LISMOD Model and Submodels The MSTRUCT Model The PATH Model The RAM Model Naming Variables and Parameters Setting Constraints on Parameters Automatic Variable Selection Estimation Criteria Relationships among Estimation Criteria Gradient, Hessian, Information Matrix, and Approximate Standard Errors Counting the Degrees of Freedom Assessment of Fit Total, Direct, and Indirect Effects Standardized Solutions Modification Indices Missing Values and the Analysis of Missing Patterns Measures of Multivariate Kurtosis Initial Estimates Use of Optimization Techniques Computational Problems Displayed Output ODS Table Names ODS Graphics -
Examples
 Estimating Covariances and Correlations Estimating Covariances and Means Simultaneously Testing Uncorrelatedness of Variables Testing Covariance Patterns Testing Some Standard Covariance Pattern Hypotheses Linear Regression Model Multivariate Regression Models Measurement Error Models Testing Specific Measurement Error Models Measurement Error Models with Multiple Predictors Measurement Error Models Specified As Linear Equations Confirmatory Factor Models Confirmatory Factor Models: Some Variations The Full Information Maximum Likelihood Method Comparing the ML and FIML Estimation Path Analysis: Stability of Alienation Simultaneous Equations with Mean Structures and Reciprocal Paths Fitting Direct Covariance Structures Confirmatory Factor Analysis: Cognitive Abilities Testing Equality of Two Covariance Matrices Using a Multiple-Group Analysis Testing Equality of Covariance and Mean Matrices between Independent Groups Illustrating Various General Modeling Languages Testing Competing Path Models for the Career Aspiration Data Fitting a Latent Growth Curve Model Higher-Order and Hierarchical Factor Models Linear Relations among Factor Loadings Multiple-Group Model for Purchasing Behavior Fitting the RAM and EQS Models by the COSAN Modeling Language Second-Order Confirmatory Factor Analysis Linear Relations among Factor Loadings: COSAN Model Specification Ordinal Relations among Factor Loadings Longitudinal Factor Analysis
Estimating Covariances and Correlations Estimating Covariances and Means Simultaneously Testing Uncorrelatedness of Variables Testing Covariance Patterns Testing Some Standard Covariance Pattern Hypotheses Linear Regression Model Multivariate Regression Models Measurement Error Models Testing Specific Measurement Error Models Measurement Error Models with Multiple Predictors Measurement Error Models Specified As Linear Equations Confirmatory Factor Models Confirmatory Factor Models: Some Variations The Full Information Maximum Likelihood Method Comparing the ML and FIML Estimation Path Analysis: Stability of Alienation Simultaneous Equations with Mean Structures and Reciprocal Paths Fitting Direct Covariance Structures Confirmatory Factor Analysis: Cognitive Abilities Testing Equality of Two Covariance Matrices Using a Multiple-Group Analysis Testing Equality of Covariance and Mean Matrices between Independent Groups Illustrating Various General Modeling Languages Testing Competing Path Models for the Career Aspiration Data Fitting a Latent Growth Curve Model Higher-Order and Hierarchical Factor Models Linear Relations among Factor Loadings Multiple-Group Model for Purchasing Behavior Fitting the RAM and EQS Models by the COSAN Modeling Language Second-Order Confirmatory Factor Analysis Linear Relations among Factor Loadings: COSAN Model Specification Ordinal Relations among Factor Loadings Longitudinal Factor Analysis - References
Example 26.24 Fitting a Latent Growth Curve Model
Latent factors in structural equation modeling are constructed to represent important unobserved hypothetical constructs. However, with some manipulations latent factors can also represent random effects in models. In this example, a simple latent growth curve model is considered. You use latent factors to represent the random intercepts and slopes in the latent growth curve model.
Sixteen individuals were invited to a training program that was designed to boost self-confidence. During the training, the individuals’ confidence levels were measured at five time points: initially and four more times separated by equal intervals. The data are stored in the following SAS data set:
data growth; input y1 y2 y3 y4 y5; datalines; 17.6 21.4 25.6 32.1 37.7 13.2 14.3 18.9 20.3 25.4 11.6 13.5 17.4 22.1 39.6 10.7 11.1 13.2 18.2 21.4 18.7 23.7 28.6 31.5 34.0 18.3 19.2 20.5 23.2 25.9 9.2 13.5 17.8 19.2 21.1 18.3 23.5 27.9 30.2 34.6 11.2 15.6 20.8 22.7 30.4 17.0 22.9 26.9 31.9 35.6 10.4 13.6 18.0 25.6 29.3 17.7 19.0 22.5 28.5 30.7 14.5 19.4 21.1 28.8 31.5 20.0 21.4 28.9 30.2 35.6 14.6 19.3 21.7 28.5 32.0 11.7 15.2 19.1 23.7 28.7 ;
First, consider a simple linear regression model for the confidence levels at time  due to training. That is,
due to training. That is,
 |
where 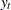 represents the confidence level at time
represents the confidence level at time  (
( ),
),  represents the intercept,
represents the intercept, 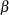 represents the slope or the effect of training,
represents the slope or the effect of training, 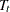 represents the fixed time point at
represents the fixed time point at  (
(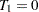 and
and 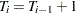 ), and
), and 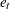 is the error term at time
is the error term at time  .
.
This simple linear regression assumes that the effect of training (slope) and the intercept are constants for the individuals. However, individual differences are rules rather than exceptions. It is thus more reasonable to argue that an index 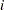 for individuals should be added to the intercept and slope in the model. As a result, the following individualized regression model is derived:
for individuals should be added to the intercept and slope in the model. As a result, the following individualized regression model is derived:
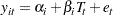 |
where 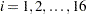 . In this model, individuals are assumed to have different intercepts and slopes (regression coefficients). Note that theoretically
. In this model, individuals are assumed to have different intercepts and slopes (regression coefficients). Note that theoretically 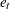 could also be "individualized" as
could also be "individualized" as 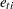 in the model. But this is not done because such a model would be unnecessarily complicated without gaining additional insights in return.
in the model. But this is not done because such a model would be unnecessarily complicated without gaining additional insights in return.
Unfortunately, this individualized model with individual intercepts and slopes cannot be estimated directly. If you treat each 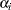 and
and 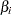 as fixed parameters, you are going to have too many parameters for the model to be identified or estimable. A workable solution is to treat
as fixed parameters, you are going to have too many parameters for the model to be identified or estimable. A workable solution is to treat  and
and 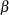 in the original linear regression model as random variables instead. That is, the latent growth curve model of interest is as follows:
in the original linear regression model as random variables instead. That is, the latent growth curve model of interest is as follows:
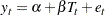 |
where 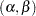 is bivariate normal with unknown means, variances, and covariance. Therefore, instead of having
is bivariate normal with unknown means, variances, and covariance. Therefore, instead of having 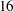 intercepts and
intercepts and 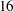 slopes to estimate in the individualized regression model, the final latent growth curve model has to estimate only two means, two variances and one covariance in the bivariate distribution of
slopes to estimate in the individualized regression model, the final latent growth curve model has to estimate only two means, two variances and one covariance in the bivariate distribution of 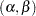 .
.
To use PROC CALIS to fit this latent growth curve model, the random intercept and effect are treated as if they were covarying latent factors. To make them stand out more as latent variables, the random intercept and slope are renamed as 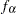 and
and 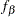 in the following structural equation:
in the following structural equation:
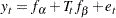 |
where 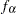 and
and 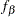 are bivariate-normal latent variables. This model assumes that the error distribution is time dependent (with the index
are bivariate-normal latent variables. This model assumes that the error distribution is time dependent (with the index  ). A simpler version is to make this error term invariant over time, which is then represented by the following model with constrained error variances:
). A simpler version is to make this error term invariant over time, which is then represented by the following model with constrained error variances:
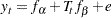 |
This constrained model is considered first. The LINEQS modeling language is used to specify this constrained model, as shown in the following statements.
proc calis method=ml data=growth nostand noparmname;
lineqs
y1 = 0. * Intercept + f_alpha + e1,
y2 = 0. * Intercept + f_alpha + 1 * f_beta + e2,
y3 = 0. * Intercept + f_alpha + 2 * f_beta + e3,
y4 = 0. * Intercept + f_alpha + 3 * f_beta + e4,
y5 = 0. * Intercept + f_alpha + 4 * f_beta + e5;
variance
f_alpha f_beta,
e1-e5 = 5 * evar;
mean
f_alpha f_beta;
cov
f_alpha f_beta;
fitindex on(only)=[chisq df probchi];
run;
In the LINEQS model specification, f_alpha and f_beta are treated as latent factors representing the random intercept and random slope, respectively. The f_ prefix for latent factors is required as a convention in the LINEQS modeling language. See the sections Naming Variables in the LINEQS Model and Naming Variables and Parameters for details.
Notice that you need to set the ordinary (non-random) intercepts for endogenous variables to zero by the 0.*Intercept specification because non-random intercepts for observed endogenous variables are default parameters in the LINEQS model. Because you have already used f_alpha as the random intercept, you must turn off the default non-random intercept term for the observed endogenous variables y1–y5. Otherwise, your latent growth curve model might be over-parameterized.
At 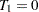 ,
, 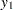 represents the initial confidence measurement so that it is not subject to the random effect f_beta. The next four measurements
represents the initial confidence measurement so that it is not subject to the random effect f_beta. The next four measurements 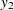 ,
, 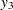 ,
, 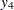 , and
, and 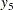 are measured at time points
are measured at time points  ,
, 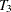 ,
, 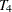 , and
, and 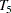 , respectively. These are fixed time points with constant values
, respectively. These are fixed time points with constant values  ,
, 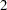 ,
, 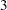 , and
, and  , respectively, in the equations of the LINEQS statement.
, respectively, in the equations of the LINEQS statement.
The means, variances and covariances of f_alpha and f_beta are parameters in the model. The variances of these two latent variables are specified in the VARIANCE statement, while their covariance is specified in the COV statement. The means of f_alpha and f_beta are specified in the MEAN statement. Unlike the specification for the variances of e1–e5. All these parameters for the latent factors are unnamed because you do not need to constrain them by references.
The error variances for e1–e5 are also specified in the VARIANCE statement. Using the shorthand notation 5 * evar, the parameter name evar is repeated five times for the five error variances. This constrains the error variances for e1–e5 to be equal.
You also use some special printing options in this example. In the PROC CALIS statement, the NOSTAND option is specified because standardized solution is not of interest. The reason is that 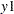 -
-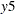 were already measured on comparable scales, making standardization unnecessary for interpretations. Another printing option specified is the NOPARMNAME option in the PROC CALIS statement. This option suppresses the printing of parameter names in the output for estimation. This makes the output look more concise when you do not need to make references to the parameter names. Still another printing option used is the ON(ONLY)= option of the FITINDEX statement. This option trims down the display of fit indices to include only those listed in the option. See the FITINDEX statement for details.
were already measured on comparable scales, making standardization unnecessary for interpretations. Another printing option specified is the NOPARMNAME option in the PROC CALIS statement. This option suppresses the printing of parameter names in the output for estimation. This makes the output look more concise when you do not need to make references to the parameter names. Still another printing option used is the ON(ONLY)= option of the FITINDEX statement. This option trims down the display of fit indices to include only those listed in the option. See the FITINDEX statement for details.
Output 26.24.1 shows the fit summary table.
| Fit Summary | |
|---|---|
| Chi-Square | 31.4310 |
| Chi-Square DF | 14 |
| Pr > Chi-Square | 0.0048 |
In Output 26.24.1, the chi-square value in the fit summary table is  (
(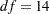 ,
, 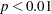 ), which is a statistically significant result that might indicate a poor model fit. Despite that, it is illustrative to continue to look at the main estimation results, which are shown in the following table.
), which is a statistically significant result that might indicate a poor model fit. Despite that, it is illustrative to continue to look at the main estimation results, which are shown in the following table.
| Estimates for Variances of Exogenous Variables | ||||
|---|---|---|---|---|
| Variable Type |
Variable | Estimate | Standard Error |
t Value |
| Latent | f_alpha | 13.89140 | 5.81540 | 2.38873 |
| f_beta | 0.80742 | 0.42198 | 1.91342 | |
| Error | e1 | 3.32185 | 0.70031 | 4.74342 |
| e2 | 3.32185 | 0.70031 | 4.74342 | |
| e3 | 3.32185 | 0.70031 | 4.74342 | |
| e4 | 3.32185 | 0.70031 | 4.74342 | |
| e5 | 3.32185 | 0.70031 | 4.74342 | |
| Covariances Among Exogenous Variables | ||||
|---|---|---|---|---|
| Var1 | Var2 | Estimate | Standard Error |
t Value |
| f_alpha | f_beta | -0.35281 | 1.13815 | -0.30998 |
| Mean Parameters | ||||
|---|---|---|---|---|
| Variable Type |
Variable | Estimate | Standard Error |
t Value |
| Latent | f_alpha | 14.15875 | 1.02906 | 13.75890 |
| f_beta | 4.04813 | 0.27563 | 14.68665 | |
In Output 26.24.2, the estimated variance of the random intercept  , which is represented by the variance estimate of the latent factor f_alpha, is
, which is represented by the variance estimate of the latent factor f_alpha, is 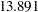 (
(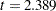 ). In the next row of the same table, the variance estimate of the random effect
). In the next row of the same table, the variance estimate of the random effect 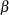 , which is represented by the variance estimate of the latent factor f_beta, is
, which is represented by the variance estimate of the latent factor f_beta, is 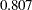 (
( ).
).
The covariance of the random intercept and the random effect is shown in the next table for "Covariances Among Exogenous Variables." A negative estimate of 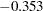 is shown. This means that the initial self-confidence level and the boosting effect of training are negatively correlated. The higher the initial self-confidence level, the smaller the training effect.
is shown. This means that the initial self-confidence level and the boosting effect of training are negatively correlated. The higher the initial self-confidence level, the smaller the training effect.
In the last table for the "Mean Parameters," the estimated mean of the random intercept is 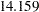 , which is an estimate of the averaged initial self-confidence level. The estimated mean of random effect is
, which is an estimate of the averaged initial self-confidence level. The estimated mean of random effect is 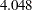 , which is an estimate of the averaged training effect. They are both significantly different from zero.
, which is an estimate of the averaged training effect. They are both significantly different from zero.
Given that the model does not fit that well, perhaps you should not take the interpretations of these estimates so seriously. Knowing that the distribution of the errors might have been time-dependent, you now try to improve the fit of the model by relaxing the constraint about common error variances. You can use the following specifications:
proc calis method=ml data=growth nostand noparmname;
lineqs
y1 = 0. * Intercept + f_alpha + e1,
y2 = 0. * Intercept + f_alpha + 1 * f_beta + e2,
y3 = 0. * Intercept + f_alpha + 2 * f_beta + e3,
y4 = 0. * Intercept + f_alpha + 3 * f_beta + e4,
y5 = 0. * Intercept + f_alpha + 4 * f_beta + e5;
variance
f_alpha f_beta,
e1-e5;
mean
f_alpha f_beta;
cov
f_alpha f_beta;
fitindex on(only)=[chisq df probchi];
run;
In this new specification, there is only one change in the VARIANCE statement from the previous specification. That is, you now specify only the error variables without putting parameter names for them. This makes the variances of e1–e5 free (unconstrained) parameters in the model.
Output 26.24.3 shows the model fit summary.
| Fit Summary | |
|---|---|
| Chi-Square | 11.6250 |
| Chi-Square DF | 10 |
| Pr > Chi-Square | 0.3109 |
The chi-square for the unconstrained model is 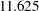 (
(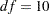 ,
, 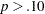 ). This indicates an acceptable model fit. The chi-square difference test can also be conducted for testing the previous constrained model against this new model. The chi-square difference is
). This indicates an acceptable model fit. The chi-square difference test can also be conducted for testing the previous constrained model against this new model. The chi-square difference is 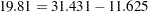 . With
. With 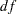 =4, this chi-square difference value is statistically significant at
=4, this chi-square difference value is statistically significant at  =0.01, indicating a significant improvement of model fit by using the unconstrained model.
=0.01, indicating a significant improvement of model fit by using the unconstrained model.
Output 26.24.4 shows the estimation results.
| Estimates for Variances of Exogenous Variables | ||||
|---|---|---|---|---|
| Variable Type |
Variable | Estimate | Standard Error |
t Value |
| Latent | f_alpha | 14.70071 | 5.66943 | 2.59298 |
| f_beta | 0.45059 | 0.29867 | 1.50867 | |
| Error | e1 | 2.81712 | 1.35332 | 2.08164 |
| e2 | 0.32213 | 0.46118 | 0.69848 | |
| e3 | 1.94429 | 0.86824 | 2.23935 | |
| e4 | 1.88569 | 1.21306 | 1.55448 | |
| e5 | 14.65193 | 5.99354 | 2.44462 | |
| Covariances Among Exogenous Variables | ||||
|---|---|---|---|---|
| Var1 | Var2 | Estimate | Standard Error |
t Value |
| f_alpha | f_beta | 0.35291 | 0.90366 | 0.39054 |
| Mean Parameters | ||||
|---|---|---|---|---|
| Variable Type |
Variable | Estimate | Standard Error |
t Value |
| Latent | f_alpha | 14.03046 | 1.01534 | 13.81851 |
| f_beta | 3.96793 | 0.22612 | 17.54781 | |
The estimation results for the unconstrained model present a slightly different picture than the constrained model. While the estimates for the means and variances of the random intercept and the random training effect look similar in both models, estimates of the covariance between the random intercept and the random training effect are quite different in the two models. The covariance estimate is negative (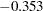 ) in the constrained model, but it is positive (
) in the constrained model, but it is positive (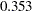 ) in the unconstrained model. However, because the covariance estimates are not statistically significant in both models (
) in the unconstrained model. However, because the covariance estimates are not statistically significant in both models (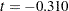 and
and 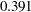 , respectively), you wonder whether the current data are showing strong evidence that supports one way or another. To get a clearer picture, perhaps you need to collect more data and fit the models again to examine the significance of the covariance between the random intercept and slope.
, respectively), you wonder whether the current data are showing strong evidence that supports one way or another. To get a clearer picture, perhaps you need to collect more data and fit the models again to examine the significance of the covariance between the random intercept and slope.