Example 26.22 Illustrating Various General Modeling Languages
In PROC CALIS, you can use many different modeling languages to specify the same model. The choice of modeling language depends on personal preferences and the purposes of the analysis. See the section Which Modeling Language? for guidance. In this example, the data and the model in Example 26.16 are used to illustrate how a particular model can be specified by various general modeling languages.
RAM Model Specification
In Example 26.16, you use the PATH modeling language to specify the model because of its close resemblance to the path diagram. In this example, you consider another modeling language of PROC CALIS that is also closely related to the path diagram representation of structural equation models. The so-called RAM model language has syntax that represents the single- and double-headed paths (or arrows) in the path diagram. However, unlike the PATH modeling language, the RAM modeling language is matrix-based. The following statements show how you can specify the same path model with the RAM model specification for the data in Example 26.16:
proc calis nobs=932 data=Wheaton;
ram
var = Anomie67 /* 1 */
Powerless67 /* 2 */
Anomie71 /* 3 */
Powerless71 /* 4 */
Education /* 5 */
SEI /* 6 */
Alien67 /* 7 */
Alien71 /* 8 */
SES, /* 9 */
_A_ 1 7 1.0,
_A_ 2 7 0.833,
_A_ 3 8 1.0,
_A_ 4 8 0.833,
_A_ 5 9 1.0,
_A_ 6 9 lambda,
_A_ 7 9 gamma1,
_A_ 8 9 gamma2,
_A_ 8 7 beta,
_P_ 1 1 theta1,
_P_ 2 2 theta2,
_P_ 3 3 theta1,
_P_ 4 4 theta2,
_P_ 5 5 theta3,
_P_ 6 6 theta4,
_P_ 7 7 psi1,
_P_ 8 8 psi2,
_P_ 9 9 phi,
_P_ 1 3 theta5,
_P_ 2 4 theta5;
run;
In the RAM model for covariance structure analysis, you have two important matrices to specify. The first one is the _A_ matrix, which is for the specification of the single-headed paths (arrows) in the path diagram. The second one is the _P_ matrix, which is for the specification of the double-headed paths (arrows) in the path diagram. Hence, to specify the RAM model is much like mapping the path diagram arrows into the parameter of the RAM model matrices.
In the RAM statement, you can specify the variables in the model in the VAR= option. The VAR= list contains all observed and latent variables in your path diagram (without the use of error terms). Although you can specify the variables in the VAR= list in any order you like, the variable order in the list is also the order of variables in the RAM model matrices. In VAR= list of the RAM statement, you put comments to note the order of the variables.
After you specify the variable list, you can specify the model parameter locations in the RAM statement entries. In the first nine entries, you specify the single-headed paths by mapping them into the elements of the _A_ matrix of the RAM model. For example, the first entry represents the single-headed path of variable 1 (Anomie67) from variable 7 (Alien67). The corresponding path effect or coefficient is fixed at 1, which is also the value for _A_[1,7]. Another example is the ninth path entry. You specify a single-headed path of variable 8 (Alien71) from variable 7 (Alien67). The corresponding path effect or coefficient is a free parameter named beta, which is also the parameter for _A_[8,7]. Hence, you can specify all single-headed paths in the path diagram as elements in the _A_ matrix of the RAM model.
To facilitate the comparisons between the RAN and PATH modeling languages, the PATH model specification in Example 26.16 for the same data is reproduced in the following:
proc calis nobs=932 data=Wheaton plots=residuals;
path
Anomie67 Powerless67 <--- Alien67 = 1.0 0.833,
Anomie71 Powerless71 <--- Alien71 = 1.0 0.833,
Education SEI <--- SES = 1.0 lambda,
Alien67 Alien71 <--- SES = gamma1 gamma2,
Alien71 <--- Alien67 = beta;
pvar
Anomie67 = theta1,
Powerless67 = theta2,
Anomie71 = theta1,
Powerless71 = theta2,
Education = theta3,
SEI = theta4,
Alien67 = psi1,
Alien71 = psi2,
SES = phi;
pcov
Anomie67 Anomie71 = theta5,
Powerless67 Powerless71 = theta5;
run;
It is clear that each of the path entries specified in the PATH statement corresponds to an matrix element entry of the _A_ matrix in the RAM statement. How about the specifications of the double-headed arrows in the path diagram? Do the RAM and PATH model specifications correspond to each other?
The answer is yes. In the PATH modeling language, you specify all double-headed arrows in the path diagram as entries either in the PVAR or PCOV statement. In the RAM modeling language, you specify the corresponding entries as matrix element entries of the _P_ matrix in the RAM statement. For example, the error variance of Anomie67 is a parameter called _Variabletheta1 in the PVAR statement of the PATH model. You specify the same parameter for the _P_[1,1] element in an entry of the RAM statement. Another example is the error covariance between Powerless67 and Powerless71. You specify this a parameter called theta5 in the last entry of the PCOV statement in the PATH model. You specify the same parameter for the _P_[2,4] element in the last entry of the RAM statement. Therefore, it is not difficult to find that the specifications in the PATH and the RAM model have some kind of one-to-one correspondence.
Output 26.22.1 shows the RAM model estimates for the Wheaton data. These RAM model estimates match the set of estimates using the PATH model specification, as shown in Output 26.16.10.
Output 26.22.1
RAM Model Estimates
| |
1.00000 |
|
|
| |
0.83300 |
|
|
| |
1.00000 |
|
|
| |
0.83300 |
|
|
| |
1.00000 |
|
|
| lambda |
5.36883 |
0.43371 |
12.37880 |
| gamma1 |
-0.62994 |
0.05634 |
-11.18092 |
| gamma2 |
-0.24086 |
0.05489 |
-4.38836 |
| beta |
0.59312 |
0.04678 |
12.67884 |
| theta1 |
3.60796 |
0.20092 |
17.95717 |
| theta2 |
3.59488 |
0.16448 |
21.85563 |
| theta1 |
3.60796 |
0.20092 |
17.95717 |
| theta2 |
3.59488 |
0.16448 |
21.85563 |
| theta3 |
2.99366 |
0.49861 |
6.00398 |
| theta4 |
259.57639 |
18.31151 |
14.17559 |
| psi1 |
5.67046 |
0.42301 |
13.40500 |
| psi2 |
4.51479 |
0.33532 |
13.46394 |
| phi |
6.61634 |
0.63914 |
10.35190 |
| theta5 |
0.90580 |
0.12167 |
7.44472 |
| theta5 |
0.90580 |
0.12167 |
7.44472 |
LINEQS Model Specification
Another way to specify the model in Example 26.16 is to use the LINEQS modeling language, which is shown in the following:
proc calis nobs=932 data=Wheaton;
lineqs
Anomie67 = 1.0 * f_Alien67 + e1,
Powerless67 = 0.833 * f_Alien67 + e2,
Anomie71 = 1.0 * f_Alien71 + e3,
Powerless71 = 0.833 * f_Alien71 + e4,
Education = 1.0 * f_SES + e5,
SEI = lambda * f_SES + e6,
f_Alien67 = gamma1 * f_SES + d1,
f_Alien71 = gamma2 * f_SES + beta * f_Alien67 + d2;
variance
E1 = theta1,
E2 = theta2,
E3 = theta1,
E4 = theta2,
E5 = theta3,
E6 = theta4,
D1 = psi1,
D2 = psi2,
f_SES = phi;
cov
E1 E3 = theta5,
E2 E4 = theta5;
run;
As compared with the PATH and RAM modeling languages, the most distinct feature of the LINEQS modeling language is the explicit use of error terms in equation specifications. In the LINEQS statement, you specify exactly one equation for each endogenous variable. In each equation, you list an endogenous variable on the left-hand-side of the equation and all its predictors on the right-hand-side of the equation. You must also include an error term in each equation. Because each endogenous variable in the LINEQS statement can only be specified in exactly one equation, the number of equations in the LINEQS model and the number of paths in the corresponding path diagram do not match necessarily. In this example, there are eight equations in the LINEQS statement, but there are nine paths in the corresponding path diagram.
In addition, in the LINEQS model, you need to follow a convention of naming latent variables. For latent variables that are neither errors nor disturbances, you must use either the 'F' or 'f' prefix. For error terms, you must use either the 'E' or 'e' prefix. For disturbances, you must use either the 'D' or 'd' prefix. However, in the PATH or RAM model specification, no such convention is imposed. For example, f_Alien67, f_Alien71, and f_SES are latent factors in the LINEQS model. They are not error terms, and so they must start with the 'f' prefix. However, this prefix is not needed in the PATH or RAM model. Furthermore, there are no explicit error terms that need to be specified in the PATH or RAM model, let alone specific prefixes for the error terms.
The PVAR statement in the PATH model is replaced with the VARIANCE statement in the LINEQS model, and the PCOV statement with the COV statement. The PVAR and PCOV statements in the PATH model are for the partial variance and partial covariance specifications. The partial variance or covariance concepts are used in the PATH or RAM model specification because error terms are not named explicitly. Specification of error variances in the PATH and RAM model is conceptualized as the specification of the partial variances of the corresponding variables. But in the LINEQS model, because errors or disturbances are named explicitly as exogenous variables, the partial variance or covariance concepts are no longer necessary. Instead, you specify the variances of the error terms directly, which reflects the conceptualization behind the VARIANCE statement of the LINEQS modeling language. Similarly, you use the COV, but not PCOV, statement in the LINEQS modeling language because you can specify the covariances among variables or error terms without using the partial covariance conceptualization.
In this example, the variances of the errors ("E"-variables) and disturbances ("D"-variables) specified in the VARIANCE statement of the LINEQS model correspond to the partial variances of the endogenous variables specified in the PVAR statement of the PATH model. Similarly, covariances of errors specified in the COV statement of the LINEQS model correspond to the partial covariances of endogenous variables specified in the PCOV statement of the PATH model. The estimation results of the LINEQS model are shown in Output 26.22.2. Again, they are essentially the same estimates obtained from the PATH model specified in Example 26.16, as shown in Output 26.16.10.
Output 26.22.2
LINEQS Model Estimates
| Anomie67 |
= |
|
1.0000 |
|
f_Alien67 |
+ |
1.0000 |
|
e1 |
|
|
|
|
| Powerless67 |
= |
|
0.8330 |
|
f_Alien67 |
+ |
1.0000 |
|
e2 |
|
|
|
|
| Anomie71 |
= |
|
1.0000 |
|
f_Alien71 |
+ |
1.0000 |
|
e3 |
|
|
|
|
| Powerless71 |
= |
|
0.8330 |
|
f_Alien71 |
+ |
1.0000 |
|
e4 |
|
|
|
|
| Education |
= |
|
1.0000 |
|
f_SES |
+ |
1.0000 |
|
e5 |
|
|
|
|
| SEI |
= |
|
5.3688 |
* |
f_SES |
+ |
1.0000 |
|
e6 |
|
|
|
|
| Std Err |
|
|
0.4337 |
|
lambda |
|
|
|
|
|
|
|
|
| t Value |
|
|
12.3788 |
|
|
|
|
|
|
|
|
|
|
| f_Alien67 |
= |
|
-0.6299 |
* |
f_SES |
+ |
1.0000 |
|
d1 |
|
|
|
|
| Std Err |
|
|
0.0563 |
|
gamma1 |
|
|
|
|
|
|
|
|
| t Value |
|
|
-11.1809 |
|
|
|
|
|
|
|
|
|
|
| f_Alien71 |
= |
|
-0.2409 |
* |
f_SES |
+ |
0.5931 |
* |
f_Alien67 |
+ |
1.0000 |
|
d2 |
| Std Err |
|
|
0.0549 |
|
gamma2 |
|
0.0468 |
|
beta |
|
|
|
|
| t Value |
|
|
-4.3884 |
|
|
|
12.6788 |
|
|
|
|
|
|
| theta1 |
3.60796 |
0.20092 |
17.95717 |
| theta2 |
3.59488 |
0.16448 |
21.85563 |
| theta1 |
3.60796 |
0.20092 |
17.95717 |
| theta2 |
3.59488 |
0.16448 |
21.85563 |
| theta3 |
2.99366 |
0.49861 |
6.00398 |
| theta4 |
259.57639 |
18.31151 |
14.17559 |
| psi1 |
5.67046 |
0.42301 |
13.40500 |
| psi2 |
4.51479 |
0.33532 |
13.46394 |
| phi |
6.61634 |
0.63914 |
10.35190 |
| theta5 |
0.90580 |
0.12167 |
7.44472 |
| theta5 |
0.90580 |
0.12167 |
7.44472 |
LISMOD Specification
You can also specify general structural models by using the LISMOD modeling language. See the section The LISMOD Model and Submodels for details.
To use the LISMOD modeling language, you must recognize four types of variables in the model. The 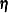 -variables (eta-variables) are latent factors that are endogenous, or predicted by other latent factors. The
-variables (eta-variables) are latent factors that are endogenous, or predicted by other latent factors. The  -variables (xi-variables) are exogenous latent variables that are not predicted by any other variables. The
-variables (xi-variables) are exogenous latent variables that are not predicted by any other variables. The 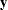 -variables are manifest variables that are indicators of the
-variables are manifest variables that are indicators of the 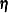 -variables, and the
-variables, and the  -variables are manifest variables that are indicators of the
-variables are manifest variables that are indicators of the  -variables. In this example, Alien67 and Alien71 are the
-variables. In this example, Alien67 and Alien71 are the 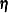 -variables, and SES is the
-variables, and SES is the  -variable in the model. Manifest indicators for Alien67 and Alien71 include Anomie67, Powerless67, Anomie71, and Powerless71, which are the
-variable in the model. Manifest indicators for Alien67 and Alien71 include Anomie67, Powerless67, Anomie71, and Powerless71, which are the 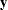 -variables. Manifest indicators for SES include Education and SEI, which are the
-variables. Manifest indicators for SES include Education and SEI, which are the  -variables.
-variables.
After defining these four types of variables, the parameters of the model are defined as entries in the model matrices. The _LAMBDAY_, _LAMBDAX_, _GAMMA_, and _BETA_ are matrices for the path coefficients or effects. The _THETAY, _THETAX_, _PSI_, and _PHI_ are matrices for the variances and covariances.
The following is the LISMOD specification for the model in Example 26.16:
proc calis nobs=932 data=Wheaton;
lismod
yvar = Anomie67 Powerless67 Anomie71 Powerless71,
xvar = Education SEI,
etavar = Alien67 Alien71,
xivar = SES;
matrix _LAMBDAY_
[1,1] = 1,
[2,1] = 0.833,
[3,2] = 1,
[4,2] = 0.833;
matrix _LAMBDAX_
[1,1] = 1,
[2,1] = lambda;
matrix _GAMMA_
[1,1] = gamma1,
[2,1] = gamma2;
matrix _BETA_
[2,1] = beta;
matrix _THETAY_
[1,1] = theta1-theta2 theta1-theta2,
[3,1] = theta5,
[4,2] = theta5;
matrix _THETAX_
[1,1] = theta3-theta4;
matrix _PSI_
[1,1] = psi1-psi2;
matrix _PHI_
[1,1] = phi;
run;
In the LISMOD statement, you specify the four lists of variables in the model. The orders of the variables in these lists define the order of the row and column variables in the model matrices, of which the parameter locations are specified in the MATRIX statements.
The estimated model is divided into three conceptual parts. The first part is the measurement model that relates the 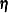 -variables with the
-variables with the 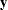 -variables, as shown in Output 26.22.3:
-variables, as shown in Output 26.22.3:
Output 26.22.3
LISMOD Model Measurement Model for the 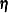 -Variables
-Variables
| 3.6080 |
| 0.2009 |
| 17.9572 |
| [theta1] |
|
|
| 0.9058 |
| 0.1217 |
| 7.4447 |
| [theta5] |
|
|
|
| 3.5949 |
| 0.1645 |
| 21.8556 |
| [theta2] |
|
|
| 0.9058 |
| 0.1217 |
| 7.4447 |
| [theta5] |
|
| 0.9058 |
| 0.1217 |
| 7.4447 |
| [theta5] |
|
|
| 3.6080 |
| 0.2009 |
| 17.9572 |
| [theta1] |
|
|
|
| 0.9058 |
| 0.1217 |
| 7.4447 |
| [theta5] |
|
|
| 3.5949 |
| 0.1645 |
| 21.8556 |
| [theta2] |
|
The _LAMBDAY_ matrix contains the coefficients or effects of the 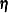 -variables on the
-variables on the 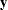 -variables. All these estimates are fixed constants as specified. The _THETAY_ matrix contains the error variances and covariances for the
-variables. All these estimates are fixed constants as specified. The _THETAY_ matrix contains the error variances and covariances for the 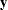 -variables. Three free parameters are located in this matrix: theta1, theta2, and theta5.
-variables. Three free parameters are located in this matrix: theta1, theta2, and theta5.
The second part of the estimated model is the measurement model that relates the  -variable with the
-variable with the  -variables, as shown in Output 26.22.4:
-variables, as shown in Output 26.22.4:
Output 26.22.4
LISMOD Model Measurement Model for the  -Variables
-Variables
|
| 5.3688 |
| 0.4337 |
| 12.3788 |
| [lambda] |
|
| 2.9937 |
| 0.4986 |
| 6.0040 |
| [theta3] |
|
|
|
| 259.5764 |
| 18.3115 |
| 14.1756 |
| [theta4] |
|
The _LAMBDAX_ matrix contains the coefficients or effects of the  -variable SES on the
-variable SES on the  -variables. The effect of SES on Education is fixed at one. The effect of SES on SEI is represented by the free parameter lambda, which is estimated at
-variables. The effect of SES on Education is fixed at one. The effect of SES on SEI is represented by the free parameter lambda, which is estimated at  . The _THETAX_ matrix contains the error variances and covariances for the
. The _THETAX_ matrix contains the error variances and covariances for the  -variables. Two free parameters are located in this matrix: theta3 and theta4.
-variables. Two free parameters are located in this matrix: theta3 and theta4.
The last part of the estimated model is the structural model that relates the latent variables 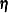 and
and  , as shown in Output 26.22.5:
, as shown in Output 26.22.5:
Output 26.22.5
LISMOD Structural Model for the Latent Variables
|
|
| 0.5931 |
| 0.0468 |
| 12.6788 |
| [beta] |
|
|
| -0.6299 |
| 0.0563 |
| -11.1809 |
| [gamma1] |
|
| -0.2409 |
| 0.0549 |
| -4.3884 |
| [gamma2] |
|
| 5.6705 |
| 0.4230 |
| 13.4050 |
| [psi1] |
|
|
|
| 4.5148 |
| 0.3353 |
| 13.4639 |
| [psi2] |
|
| 6.6163 |
| 0.6391 |
| 10.3519 |
| [phi] |
|
The _BETA_ matrix contains effects of 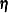 -variables on themselves. In the current example, there is only one such effect. The effect of Alien67 on Alien71 is represented by the free parameter beta. The _GAMMA_ matrix contains effects of the
-variables on themselves. In the current example, there is only one such effect. The effect of Alien67 on Alien71 is represented by the free parameter beta. The _GAMMA_ matrix contains effects of the  -variable, which is SES in this example, on the
-variable, which is SES in this example, on the 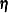 -variables Alien67 on Alien71. These effects are represented by the free parameters gamma1 and gamma2. The _PSI_ matrix contains the error variances and covariances in the structural model. In this example, psi1 and psi2 are two free parameters for the error variances. Finally, the _PHI_ matrix is the covariance matrix for the
-variables Alien67 on Alien71. These effects are represented by the free parameters gamma1 and gamma2. The _PSI_ matrix contains the error variances and covariances in the structural model. In this example, psi1 and psi2 are two free parameters for the error variances. Finally, the _PHI_ matrix is the covariance matrix for the  -variables. In this example, there is only one
-variables. In this example, there is only one  -variable so that this matrix contains only the estimated variance of SES. This variance is represented by the parameter phi.
-variable so that this matrix contains only the estimated variance of SES. This variance is represented by the parameter phi.
The estimates obtained from fitting the LISMOD model are the same as those from fitting the equivalent PATH, RAM, or LINEQS model. To some researchers the LISMOD modeling language might be more familiar, while for others modeling languages such as PATH, RAM, or LINEQS are more convenient to use.


 Classes of Statements in PROC CALIS Single-Group Analysis Syntax Multiple-Group Multiple-Model Analysis Syntax PROC CALIS Statement BOUNDS Statement BY Statement COSAN Statement COV Statement DETERM Statement EFFPART Statement FACTOR Statement FITINDEX Statement FREQ Statement GROUP Statement LINCON Statement LINEQS Statement LISMOD Statement LMTESTS Statement MATRIX Statement MEAN Statement MODEL Statement MSTRUCT Statement NLINCON Statement NLOPTIONS Statement OUTFILES Statement PARAMETERS Statement PARTIAL Statement PATH Statement PCOV Statement PVAR Statement RAM Statement REFMODEL Statement RENAMEPARM Statement SAS Programming Statements SIMTESTS Statement STD Statement STRUCTEQ Statement TESTFUNC Statement VAR Statement VARIANCE Statement VARNAMES Statement WEIGHT Statement
Classes of Statements in PROC CALIS Single-Group Analysis Syntax Multiple-Group Multiple-Model Analysis Syntax PROC CALIS Statement BOUNDS Statement BY Statement COSAN Statement COV Statement DETERM Statement EFFPART Statement FACTOR Statement FITINDEX Statement FREQ Statement GROUP Statement LINCON Statement LINEQS Statement LISMOD Statement LMTESTS Statement MATRIX Statement MEAN Statement MODEL Statement MSTRUCT Statement NLINCON Statement NLOPTIONS Statement OUTFILES Statement PARAMETERS Statement PARTIAL Statement PATH Statement PCOV Statement PVAR Statement RAM Statement REFMODEL Statement RENAMEPARM Statement SAS Programming Statements SIMTESTS Statement STD Statement STRUCTEQ Statement TESTFUNC Statement VAR Statement VARIANCE Statement VARNAMES Statement WEIGHT Statement Input Data Sets Output Data Sets The COSAN Model The FACTOR Model The LINEQS Model The LISMOD Model and Submodels The MSTRUCT Model The PATH Model The RAM Model Naming Variables and Parameters Setting Constraints on Parameters Automatic Variable Selection Estimation Criteria Relationships among Estimation Criteria Gradient, Hessian, Information Matrix, and Approximate Standard Errors Counting the Degrees of Freedom Assessment of Fit Total, Direct, and Indirect Effects Standardized Solutions Modification Indices Missing Values and the Analysis of Missing Patterns Measures of Multivariate Kurtosis Initial Estimates Use of Optimization Techniques Computational Problems Displayed Output ODS Table Names ODS Graphics
Input Data Sets Output Data Sets The COSAN Model The FACTOR Model The LINEQS Model The LISMOD Model and Submodels The MSTRUCT Model The PATH Model The RAM Model Naming Variables and Parameters Setting Constraints on Parameters Automatic Variable Selection Estimation Criteria Relationships among Estimation Criteria Gradient, Hessian, Information Matrix, and Approximate Standard Errors Counting the Degrees of Freedom Assessment of Fit Total, Direct, and Indirect Effects Standardized Solutions Modification Indices Missing Values and the Analysis of Missing Patterns Measures of Multivariate Kurtosis Initial Estimates Use of Optimization Techniques Computational Problems Displayed Output ODS Table Names ODS Graphics Estimating Covariances and Correlations Estimating Covariances and Means Simultaneously Testing Uncorrelatedness of Variables Testing Covariance Patterns Testing Some Standard Covariance Pattern Hypotheses Linear Regression Model Multivariate Regression Models Measurement Error Models Testing Specific Measurement Error Models Measurement Error Models with Multiple Predictors Measurement Error Models Specified As Linear Equations Confirmatory Factor Models Confirmatory Factor Models: Some Variations The Full Information Maximum Likelihood Method Comparing the ML and FIML Estimation Path Analysis: Stability of Alienation Simultaneous Equations with Mean Structures and Reciprocal Paths Fitting Direct Covariance Structures Confirmatory Factor Analysis: Cognitive Abilities Testing Equality of Two Covariance Matrices Using a Multiple-Group Analysis Testing Equality of Covariance and Mean Matrices between Independent Groups Illustrating Various General Modeling Languages Testing Competing Path Models for the Career Aspiration Data Fitting a Latent Growth Curve Model Higher-Order and Hierarchical Factor Models Linear Relations among Factor Loadings Multiple-Group Model for Purchasing Behavior Fitting the RAM and EQS Models by the COSAN Modeling Language Second-Order Confirmatory Factor Analysis Linear Relations among Factor Loadings: COSAN Model Specification Ordinal Relations among Factor Loadings Longitudinal Factor Analysis
Estimating Covariances and Correlations Estimating Covariances and Means Simultaneously Testing Uncorrelatedness of Variables Testing Covariance Patterns Testing Some Standard Covariance Pattern Hypotheses Linear Regression Model Multivariate Regression Models Measurement Error Models Testing Specific Measurement Error Models Measurement Error Models with Multiple Predictors Measurement Error Models Specified As Linear Equations Confirmatory Factor Models Confirmatory Factor Models: Some Variations The Full Information Maximum Likelihood Method Comparing the ML and FIML Estimation Path Analysis: Stability of Alienation Simultaneous Equations with Mean Structures and Reciprocal Paths Fitting Direct Covariance Structures Confirmatory Factor Analysis: Cognitive Abilities Testing Equality of Two Covariance Matrices Using a Multiple-Group Analysis Testing Equality of Covariance and Mean Matrices between Independent Groups Illustrating Various General Modeling Languages Testing Competing Path Models for the Career Aspiration Data Fitting a Latent Growth Curve Model Higher-Order and Hierarchical Factor Models Linear Relations among Factor Loadings Multiple-Group Model for Purchasing Behavior Fitting the RAM and EQS Models by the COSAN Modeling Language Second-Order Confirmatory Factor Analysis Linear Relations among Factor Loadings: COSAN Model Specification Ordinal Relations among Factor Loadings Longitudinal Factor Analysis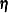 -variables (eta-variables) are latent factors that are endogenous, or predicted by other latent factors. The
-variables (eta-variables) are latent factors that are endogenous, or predicted by other latent factors. The  -variables (xi-variables) are exogenous latent variables that are not predicted by any other variables. The
-variables (xi-variables) are exogenous latent variables that are not predicted by any other variables. The 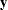 -variables are manifest variables that are indicators of the
-variables are manifest variables that are indicators of the 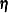 -variables, and the
-variables, and the  -variables are manifest variables that are indicators of the
-variables are manifest variables that are indicators of the  -variables. In this example, Alien67 and Alien71 are the
-variables. In this example, Alien67 and Alien71 are the 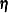 -variables, and SES is the
-variables, and SES is the  -variable in the model. Manifest indicators for Alien67 and Alien71 include Anomie67, Powerless67, Anomie71, and Powerless71, which are the
-variable in the model. Manifest indicators for Alien67 and Alien71 include Anomie67, Powerless67, Anomie71, and Powerless71, which are the 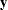 -variables. Manifest indicators for SES include Education and SEI, which are the
-variables. Manifest indicators for SES include Education and SEI, which are the  -variables.
-variables. 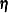 -variables with the
-variables with the 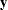 -variables, as shown in Output 26.22.3:
-variables, as shown in Output 26.22.3: 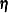 -Variables
-Variables
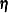 -variables on the
-variables on the 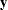 -variables. All these estimates are fixed constants as specified. The _THETAY_ matrix contains the error variances and covariances for the
-variables. All these estimates are fixed constants as specified. The _THETAY_ matrix contains the error variances and covariances for the 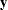 -variables. Three free parameters are located in this matrix: theta1, theta2, and theta5.
-variables. Three free parameters are located in this matrix: theta1, theta2, and theta5.  -variable with the
-variable with the  -variables, as shown in Output 26.22.4:
-variables, as shown in Output 26.22.4:  -Variables
-Variables
 -variable SES on the
-variable SES on the  -variables. The effect of SES on Education is fixed at one. The effect of SES on SEI is represented by the free parameter lambda, which is estimated at
-variables. The effect of SES on Education is fixed at one. The effect of SES on SEI is represented by the free parameter lambda, which is estimated at  . The _THETAX_ matrix contains the error variances and covariances for the
. The _THETAX_ matrix contains the error variances and covariances for the  -variables. Two free parameters are located in this matrix: theta3 and theta4.
-variables. Two free parameters are located in this matrix: theta3 and theta4. 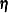 and
and  , as shown in Output 26.22.5:
, as shown in Output 26.22.5: 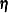 -variables on themselves. In the current example, there is only one such effect. The effect of Alien67 on Alien71 is represented by the free parameter beta. The _GAMMA_ matrix contains effects of the
-variables on themselves. In the current example, there is only one such effect. The effect of Alien67 on Alien71 is represented by the free parameter beta. The _GAMMA_ matrix contains effects of the  -variable, which is SES in this example, on the
-variable, which is SES in this example, on the 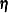 -variables Alien67 on Alien71. These effects are represented by the free parameters gamma1 and gamma2. The _PSI_ matrix contains the error variances and covariances in the structural model. In this example, psi1 and psi2 are two free parameters for the error variances. Finally, the _PHI_ matrix is the covariance matrix for the
-variables Alien67 on Alien71. These effects are represented by the free parameters gamma1 and gamma2. The _PSI_ matrix contains the error variances and covariances in the structural model. In this example, psi1 and psi2 are two free parameters for the error variances. Finally, the _PHI_ matrix is the covariance matrix for the  -variables. In this example, there is only one
-variables. In this example, there is only one  -variable so that this matrix contains only the estimated variance of SES. This variance is represented by the parameter phi.
-variable so that this matrix contains only the estimated variance of SES. This variance is represented by the parameter phi.