The MIXED Procedure
Clustered Data Example
Consider the following SAS data set as an introductory example:
data heights; input Family Gender$ Height @@; datalines; 1 F 67 1 F 66 1 F 64 1 M 71 1 M 72 2 F 63 2 F 63 2 F 67 2 M 69 2 M 68 2 M 70 3 F 63 3 M 64 4 F 67 4 F 66 4 M 67 4 M 67 4 M 69 ;
The response variable Height measures the heights (in inches) of 18 individuals. The individuals are classified according to Family and Gender. You can perform a traditional two-way analysis of variance of these data with the following PROC MIXED statements:
proc mixed data=heights; class Family Gender; model Height = Gender Family Family*Gender; run;
The PROC MIXED
statement invokes the procedure. The CLASS
statement instructs PROC MIXED to consider both Family and Gender as classification variables. Dummy (indicator) variables are, as a result, created corresponding to all of the distinct levels
of Family and Gender. For these data, Family has four levels and Gender has two levels.
The MODEL
statement first specifies the response (dependent) variable Height. The explanatory (independent) variables are then listed after the equal (=) sign. Here, the two explanatory variables are
Gender and Family, and these are the main effects of the design. The third explanatory term, Family*Gender, models an interaction between the two main effects.
PROC MIXED uses the dummy variables associated with Gender, Family, and Family*Gender to construct the 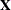 matrix for the linear model. A column of 1s is also included as the first column of
matrix for the linear model. A column of 1s is also included as the first column of 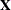 to model a global intercept. There are no
to model a global intercept. There are no 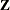 or
or 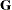 matrices for this model, and
matrices for this model, and  is assumed to equal
is assumed to equal 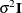 , where
, where 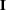 is an
is an 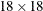 identity matrix.
identity matrix.
The RUN statement completes the specification. The coding is precisely the same as with the GLM procedure. However, much of the output from PROC MIXED is different from that produced by PROC GLM.
The output from PROC MIXED is shown in Figure 77.1–Figure 77.7.
The "Model Information" table in Figure 77.1 describes the model, some of the variables that it involves, and the method used in fitting it. This table also lists the method (profile, factor, parameter, or none) for handling the residual variance.
Figure 77.1: Model Information
The "Class Level Information" table in Figure 77.2 lists the levels of all variables specified in the CLASS statement. You can check this table to make sure that the data are correct.
Figure 77.2: Class Level Information
The "Dimensions" table in Figure 77.3 lists the sizes of relevant matrices. This table can be useful in determining CPU time and memory requirements.
Figure 77.3: Dimensions
The "Number of Observations" table in Figure 77.4 displays information about the sample size being processed.
Figure 77.4: Number of Observations
The "Covariance Parameter Estimates" table in Figure 77.5 displays the estimate of 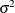 for the model.
for the model.
Figure 77.5: Covariance Parameter Estimates
The "Fit Statistics" table in Figure 77.6 lists several pieces of information about the fitted mixed model, including values derived from the computed value of the restricted/residual likelihood.
Figure 77.6: Fit Statistics
The "Type 3 Tests of Fixed Effects" table in Figure 77.7 displays significance tests for the three effects listed in the MODEL statement. The Type 3 F statistics and p-values are the same as those produced by the GLM procedure. However, because PROC MIXED uses a likelihood-based estimation scheme, it does not directly compute or display sums of squares for this analysis.
Figure 77.7: Tests of Fixed Effects
The Type 3 test for Family*Gender effect is not significant at the 5% level, but the tests for both main effects are significant.
The important assumptions behind this analysis are that the data are normally distributed and that they are independent with constant variance. For these data, the normality assumption is probably realistic since the data are observed heights. However, since the data occur in clusters (families), it is very likely that observations from the same family are statistically correlated—that is, not independent.
The methods implemented in PROC MIXED are still based on the assumption of normally distributed data, but you can drop the assumption of independence by modeling statistical correlation in a variety of ways. You can also model variances that are heterogeneous—that is, nonconstant.
For the height data, one of the simplest ways of modeling correlation is through the use of random effects. Here the family effect is assumed to be normally distributed with zero mean and some unknown variance. This is in contrast
to the previous model in which the family effects are just constants, or fixed effects. Declaring Family as a random effect sets up a common correlation among all observations having the same level of Family.
Declaring Family*Gender as a random effect models an additional correlation between all observations that have the same level of both Family and Gender. One interpretation of this effect is that a female in a certain family exhibits more correlation with the other females
in that family than with the other males, and likewise for a male. With the height data, this model seems reasonable.
The statements to fit this correlation model in PROC MIXED are as follows:
proc mixed; class Family Gender; model Height = Gender; random Family Family*Gender; run;
Note that Family and Family*Gender are now listed in the RANDOM
statement. The dummy variables associated with them are used to construct the 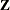 matrix in the mixed model. The
matrix in the mixed model. The 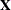 matrix now consists of a column of 1s and the dummy variables for
matrix now consists of a column of 1s and the dummy variables for Gender.
The 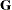 matrix for this model is diagonal, and it contains the variance components for both
matrix for this model is diagonal, and it contains the variance components for both Family and Family*Gender. The  matrix is still assumed to equal
matrix is still assumed to equal 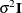 , where
, where 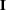 is an identity matrix.
is an identity matrix.
The output from this analysis is as follows.
Figure 77.8: Model Information
The "Model Information" table in Figure 77.8 shows that the containment method is used to compute the degrees of freedom for this analysis. This is the default method when a RANDOM statement is used; for more information, see the description of the DDFM= option.
Figure 77.9: Class Level Information
The "Class Level Information" table in Figure 77.9 is the same as before. The "Dimensions" table in Figure 77.10 displays the new sizes of the 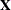 and
and 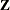 matrices.
matrices.
Figure 77.10: Dimensions and Number of Observations
The "Iteration History" table in Figure 77.11 displays the results of the numerical optimization of the restricted/residual likelihood. Six iterations are required to achieve the default convergence criterion of 1E–8.
Figure 77.11: REML Estimation Iteration History
The "Covariance Parameter Estimates" table in Figure 77.12 displays the results of the REML fit. The Estimate column contains the estimates of the variance components for Family and Family*Gender, as well as the estimate of 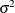 .
.
Figure 77.12: Covariance Parameter Estimates (REML)
The "Fit Statistics" table in Figure 77.13 contains basic information about the REML fit.
Figure 77.13: Fit Statistics
The "Type 3 Tests of Fixed Effects" table in Figure 77.14 contains a significance test for the lone fixed effect, Gender. Note that the associated p-value is not nearly as significant as in the previous analysis. This illustrates the importance of correctly modeling correlation
in your data.
Figure 77.14: Type 3 Tests of Fixed Effects
An additional benefit of the random effects analysis is that it enables you to make inferences about gender that apply to
an entire population of families, whereas the inferences about gender from the analysis where Family and Family*Gender are fixed effects apply only to the particular families in the data set.
PROC MIXED thus offers you the ability to model correlation directly and to make inferences about fixed effects that apply to entire populations of random effects.
