The MIXED Procedure
Residuals and Influence Diagnostics
Residual Diagnostics
Consider a residual vector of the form 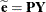 , where
, where 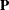 is a projection matrix, possibly an oblique projector. A typical element
is a projection matrix, possibly an oblique projector. A typical element 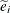 with variance
with variance 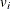 and estimated variance
and estimated variance 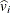 is said to be standardized as
is said to be standardized as
![\[ \frac{\widetilde{e}_ i}{\sqrt {\mr{Var}[\widetilde{e}_ i]}} = \frac{\widetilde{e}_ i}{\sqrt {v_ i}} \]](images/statug_mixed0414.png)
and studentized as
![\[ \frac{\widetilde{e}_ i}{\sqrt {\widehat{v}_ i}} \]](images/statug_mixed0415.png)
External studentization uses an estimate of ![$\mr{Var}[\widetilde{e}_ i]$](images/statug_mixed0416.png) that does not involve the ith observation. Externally studentized residuals are often preferred over internally studentized residuals because they have
well-known distributional properties in standard linear models for independent data.
that does not involve the ith observation. Externally studentized residuals are often preferred over internally studentized residuals because they have
well-known distributional properties in standard linear models for independent data.
Residuals that are scaled by the estimated variance of the response, i.e., ![$\widetilde{e}_ i/\sqrt {\widehat{\mr{Var}}[Y_ i]}$](images/statug_mixed0417.png) , are referred to as Pearson-type residuals.
, are referred to as Pearson-type residuals.
Marginal and Conditional Residuals
The marginal and conditional means in the linear mixed model are ![$\textup{E}[\mb{Y}] = \mb{X}\bbeta $](images/statug_mixed0418.png) and
and ![$\textup{E}[\mb{Y} | \bgamma ] = \mb{X}\bbeta + \mb{Z}\bgamma $](images/statug_mixed0419.png) , respectively. Accordingly, the vector
, respectively. Accordingly, the vector 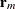 of marginal residuals is defined as
of marginal residuals is defined as
![\[ \mb{r}_ m = \mb{Y} - \mb{X}\widehat{\bbeta } \]](images/statug_mixed0421.png)
and the vector 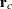 of conditional residuals is
of conditional residuals is
![\[ \mb{r}_ c = \mb{Y} - \mb{X}\widehat{\bbeta } - \mb{Z}\widehat{\bgamma } = \mb{r}_ m - \mb{Z}\widehat{\bgamma } \]](images/statug_mixed0423.png)
Following Gregoire, Schabenberger, and Barrett (1995), let 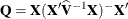 and
and 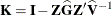 . Then
. Then
![\begin{align*} \widehat{\mr{Var}}[\mb{r}_ m] & = \widehat{\mb{V}} - \mb{Q} \\ \widehat{\mr{Var}}[\mb{r}_ c] & = \mb{K}(\widehat{\mb{V}} - \mb{Q})\mb{K}’\\ \end{align*}](images/statug_mixed0426.png)
For an individual observation the raw, studentized, and Pearson-type residuals computed by the MIXED procedure are given in Table 77.25.
Table 77.25: Residual Types Computed by the MIXED Procedure
|
Type of Residual |
Marginal |
Conditional |
|---|---|---|
|
Raw |
|
|
|
Studentized |
|
|
|
Pearson |
|
|
When the OUTPM=
option is specified in addition to the RESIDUAL
option in the MODEL
statement, 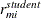 and
and 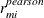 are added to the data set as variables
are added to the data set as variables Resid, StudentResid, and PearsonResid, respectively. When the OUTP=
option is specified, 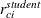 and
and 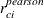 are added to the data set. Raw residuals are part of the OUTPM=
and OUTP=
data sets without the RESIDUAL
option.
are added to the data set. Raw residuals are part of the OUTPM=
and OUTP=
data sets without the RESIDUAL
option.
Scaled Residuals
For correlated data, a set of scaled quantities can be defined through the Cholesky decomposition of the variance-covariance
matrix. Since fitted residuals in linear models are rank-deficient, it is customary to draw on the variance-covariance matrix
of the data. If ![$\mr{Var}[\mb{Y}] = \bV $](images/statug_mixed0437.png) and
and 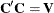 , then
, then 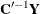 has uniform dispersion and its elements are uncorrelated.
has uniform dispersion and its elements are uncorrelated.
Scaled residuals in a mixed model are meaningful for quantities based on the marginal distribution of the data. Let 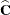 denote the Cholesky root of
denote the Cholesky root of 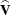 , so that
, so that 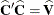 , and define
, and define
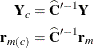
By analogy with other scalings, the inverse Cholesky decomposition can also be applied to the residual vector, 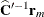 , although
, although 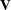 is not the variance-covariance matrix of
is not the variance-covariance matrix of 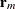 .
.
To diagnose whether the covariance structure of the model has been specified correctly can be difficult based on 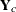 , since the inverse Cholesky transformation affects the expected value of
, since the inverse Cholesky transformation affects the expected value of 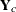 . You can draw on
. You can draw on 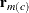 as a vector of (approximately) uncorrelated data with constant mean.
as a vector of (approximately) uncorrelated data with constant mean.
When the OUTPM=
option in the MODEL
statement is specified in addition to the VCIRY
option, 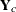 is added as variable
is added as variable ScaledDep and 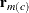 is added as
is added as ScaledResid to the data set.
Influence Diagnostics
Basic Idea and Statistics
The general idea of quantifying the influence of one or more observations relies on computing parameter estimates based on all data points, removing the cases in question from the data, refitting the model, and computing statistics based on the change between full-data and reduced-data estimation. Influence statistics can be coarsely grouped by the aspect of estimation that is their primary target:
-
overall measures compare changes in objective functions: (restricted) likelihood distance (Cook and Weisberg 1982, Ch. 5.2)
-
influence on parameter estimates: Cook’s D (Cook 1977, 1979), MDFFITS (Belsley, Kuh, and Welsch 1980, p. 32)
-
influence on precision of estimates: CovRatio and CovTrace
-
influence on fitted and predicted values: PRESS residual, PRESS statistic (Allen 1974), DFFITS (Belsley, Kuh, and Welsch 1980, p. 15)
-
outlier properties: internally and externally studentized residuals, leverage
For linear models for uncorrelated data, it is not necessary to refit the model after removing a data point in order to measure the impact of an observation on the model. The change in fixed effect estimates, residuals, residual sums of squares, and the variance-covariance matrix of the fixed effects can be computed based on the fit to the full data alone. By contrast, in mixed models several important complications arise. Data points can affect not only the fixed effects but also the covariance parameter estimates on which the fixed-effects estimates depend. Furthermore, closed-form expressions for computing the change in important model quantities might not be available.
This section provides background material for the various influence diagnostics available with the MIXED procedure. See the
section Mixed Models Theory for relevant expressions and definitions. The parameter vector 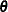 denotes all unknown parameters in the
denotes all unknown parameters in the  and
and 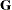 matrix.
matrix.
The observations whose influence is being ascertained are represented by the set U and referred to simply as "the observations in U." The estimate of a parameter vector, such as 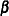 , obtained from all observations except those in the set U is denoted
, obtained from all observations except those in the set U is denoted 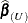 . In case of a matrix
. In case of a matrix 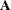 , the notation
, the notation 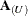 represents the matrix with the rows in U removed; these rows are collected in
represents the matrix with the rows in U removed; these rows are collected in 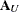 . If
. If 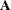 is symmetric, then notation
is symmetric, then notation 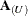 implies removal of rows and columns. The vector
implies removal of rows and columns. The vector 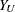 comprises the responses of the data points being removed, and
comprises the responses of the data points being removed, and 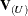 is the variance-covariance matrix of the remaining observations. When k = 1, lowercase notation emphasizes that single points are removed, such as
is the variance-covariance matrix of the remaining observations. When k = 1, lowercase notation emphasizes that single points are removed, such as 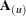 .
.
Managing the Covariance Parameters
An important component of influence diagnostics in the mixed model is the estimated variance-covariance matrix 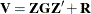 . To make the dependence on the vector of covariance parameters explicit, write it as
. To make the dependence on the vector of covariance parameters explicit, write it as 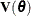 . If one parameter,
. If one parameter, 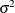 , is profiled or factored out of
, is profiled or factored out of 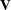 , the remaining parameters are denoted as
, the remaining parameters are denoted as 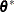 . Notice that in a model where
. Notice that in a model where 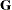 is diagonal and
is diagonal and 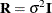 , the parameter vector
, the parameter vector 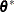 contains the ratios of each variance component and
contains the ratios of each variance component and 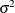 (see Wolfinger, Tobias, and Sall 1994). When ITER=
0, two scenarios are distinguished:
(see Wolfinger, Tobias, and Sall 1994). When ITER=
0, two scenarios are distinguished:
-
If the residual variance is not profiled, either because the model does not contain a residual variance or because it is part of the Newton-Raphson iterations, then
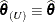 .
.
-
If the residual variance is profiled, then
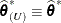 and
and 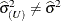 . Influence statistics such as Cook’s D and internally studentized residuals are based on
. Influence statistics such as Cook’s D and internally studentized residuals are based on 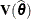 , whereas externally studentized residuals and the DFFITS statistic are based on
, whereas externally studentized residuals and the DFFITS statistic are based on 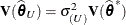 . In a random components model with uncorrelated errors, for example, the computation of
. In a random components model with uncorrelated errors, for example, the computation of 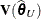 involves scaling of
involves scaling of 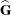 and
and 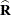 by the full-data estimate
by the full-data estimate 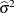 and multiplying the result with the reduced-data estimate
and multiplying the result with the reduced-data estimate 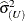 .
.
Certain statistics, such as MDFFITS, CovRatio, and CovTrace, require an estimate of the variance of the fixed effects that
is based on the reduced number of observations. For example, 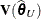 is evaluated at the reduced-data parameter estimates but computed for the entire data set. The matrix
is evaluated at the reduced-data parameter estimates but computed for the entire data set. The matrix 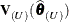 , on the other hand, has rows and columns corresponding to the points in U removed. The resulting matrix is evaluated at the delete-case estimates.
, on the other hand, has rows and columns corresponding to the points in U removed. The resulting matrix is evaluated at the delete-case estimates.
When influence analysis is iterative, the entire vector 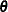 is updated, whether the residual variance is profiled or not. The matrices to be distinguished here are
is updated, whether the residual variance is profiled or not. The matrices to be distinguished here are 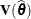 ,
, 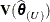 , and
, and 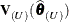 , with unambiguous notation.
, with unambiguous notation.
Predicted Values, PRESS Residual, and PRESS Statistic
An unconditional predicted value is 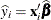 , where the vector
, where the vector 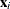 is the ith row of
is the ith row of 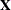 . The (raw) residual is given as
. The (raw) residual is given as 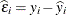 , and the PRESS residual is
, and the PRESS residual is
![\[ \widehat{\epsilon }_{i(U)} = y_ i - \mb{x}_ i’\widehat{\bbeta }_{(U)} \]](images/statug_mixed0471.png)
The PRESS statistic is the sum of the squared PRESS residuals,
![\[ \mathit{PRESS} = \sum _{i \in U} \widehat{\epsilon }_{i(U)}^{\, 2} \]](images/statug_mixed0472.png)
where the sum is over the observations in U.
If EFFECT= , SIZE= , or KEEP= is not specified, PROC MIXED computes the PRESS residual for each observation selected through SELECT= (or all observations if SELECT= is not given). If EFFECT= , SIZE= , or KEEP= is specified, the procedure computes PRESS.
Leverage
For the general mixed model, leverage can be defined through the projection matrix that results from a transformation of the
model with the inverse of the Cholesky decomposition of 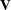 , or through an oblique projector. The MIXED procedure follows the latter path in the computation of influence diagnostics.
The leverage value reported for the ith observation is the ith diagonal entry of the matrix
, or through an oblique projector. The MIXED procedure follows the latter path in the computation of influence diagnostics.
The leverage value reported for the ith observation is the ith diagonal entry of the matrix
![\[ \bH = \mb{X}(\mb{X}’\mb{V}(\widehat{\btheta })^{-1}\mb{X})^{-} \mb{X}’\mb{V}(\widehat{\btheta })^{-1} \]](images/statug_mixed0473.png)
which is the weight of the observation in contributing to its own predicted value, 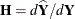 .
.
While 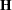 is idempotent, it is generally not symmetric and thus not a projection matrix in the narrow sense.
is idempotent, it is generally not symmetric and thus not a projection matrix in the narrow sense.
The properties of these leverages are generalizations of the properties in models with diagonal variance-covariance matrices.
For example, 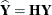 , and in a model with intercept and
, and in a model with intercept and 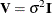 , the leverage values
, the leverage values
![\[ h_{ii} = \mb{x}_ i’(\mb{X}’\mb{X})^{-}\mb{x}_ i \]](images/statug_mixed0478.png)
are 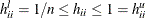 and
and 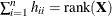 . The lower bound for
. The lower bound for 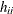 is achieved in an intercept-only model, and the upper bound is achieved in a saturated model.
The trace of
is achieved in an intercept-only model, and the upper bound is achieved in a saturated model.
The trace of 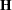 equals the rank of
equals the rank of 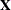 .
.
If 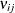 denotes the element in row i, column j of
denotes the element in row i, column j of 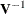 , then for a model containing only an intercept the diagonal elements of
, then for a model containing only an intercept the diagonal elements of 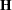 are
are
![\[ h_{ii} = \frac{\sum _{j=1}^{n} \nu _{ij}}{\sum _{i=1}^{n}\, \sum _{j=1}^{n} \nu _{ij}} \]](images/statug_mixed0484.png)
Because 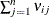 is a sum of elements in the ith row of the inverse variance-covariance matrix,
is a sum of elements in the ith row of the inverse variance-covariance matrix, 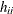 can be negative, even if the correlations among data points are nonnegative. In case of a saturated model with
can be negative, even if the correlations among data points are nonnegative. In case of a saturated model with 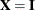 ,
, 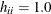 .
.
Internally and Externally Studentized Residuals
See the section Residual Diagnostics for the distinction between standardization, studentization, and scaling of residuals. Internally studentized marginal and conditional residuals are computed with the RESIDUAL option of the MODEL statement. The INFLUENCE option computes internally and externally studentized marginal residuals.
The computation of internally studentized residuals relies on the diagonal entries of 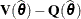 , where
, where 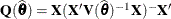 . Externally studentized residuals require iterative influence analysis or a profiled residual variance. In the former case
the studentization is based on
. Externally studentized residuals require iterative influence analysis or a profiled residual variance. In the former case
the studentization is based on 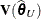 ; in the latter case it is based on
; in the latter case it is based on 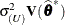 .
.
Cook’s D
Cook’s D statistic is an invariant norm that measures the influence of observations in U on a vector of parameter estimates (Cook 1977). In case of the fixed-effects coefficients, let
![\[ \bdelta _{(U)} = \widehat{\bbeta } - \widehat{\bbeta }_{(U)} \]](images/statug_mixed0491.png)
Then the MIXED procedure computes
![\[ D(\bbeta ) = \bdelta _{(U)}’ \widehat{\mr{Var}}[\widehat{\bbeta }]^{-} \bdelta _{(U)} / \mr{rank}(\mb{X}) \]](images/statug_mixed0492.png)
where ![$\widehat{\mr{Var}}[\widehat{\bbeta }]^{-}$](images/statug_mixed0493.png) is the
matrix that results from sweeping
is the
matrix that results from sweeping 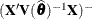 .
.
If 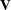 is known, Cook’s D can be calibrated according to a chi-square distribution with degrees of freedom equal to the rank of
is known, Cook’s D can be calibrated according to a chi-square distribution with degrees of freedom equal to the rank of 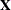 (Christensen, Pearson, and Johnson 1992). For estimated
(Christensen, Pearson, and Johnson 1992). For estimated 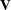 the calibration can be carried out according to an
the calibration can be carried out according to an 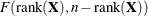 distribution. To interpret D on a familiar scale, Cook (1979) and Cook and Weisberg (1982, p. 116) refer to the 50th percentile of the reference distribution. If D is equal to that percentile, then removing the points in U moves the fixed-effects coefficient vector from the center of the confidence region to the 50% confidence ellipsoid (Myers
1990, p. 262).
distribution. To interpret D on a familiar scale, Cook (1979) and Cook and Weisberg (1982, p. 116) refer to the 50th percentile of the reference distribution. If D is equal to that percentile, then removing the points in U moves the fixed-effects coefficient vector from the center of the confidence region to the 50% confidence ellipsoid (Myers
1990, p. 262).
In the case of iterative influence analysis, the MIXED procedure also computes a D-type statistic for the covariance parameters. If  is the asymptotic variance-covariance matrix of
is the asymptotic variance-covariance matrix of 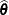 , then MIXED computes
, then MIXED computes
![\[ D_{\btheta } = (\widehat{\btheta } - \widehat{\btheta }_{(U))})’ \widehat{\bGamma }^{-1} (\widehat{\btheta } - \widehat{\btheta }_{(U)}) \]](images/statug_mixed0497.png)
DFFITS and MDFFITS
A DFFIT measures the change in predicted values due to removal of data points. If this change is standardized by the externally estimated standard error of the predicted value in the full data, the DFFITS statistic of Belsley, Kuh, and Welsch (1980, p. 15) results:
![\[ \mr{DFFITS}_ i = (\widehat{y}_ i - \widehat{y}_{i(u)} ) / \mr{ese}(\widehat{y}_ i) \]](images/statug_mixed0498.png)
The MIXED procedure computes DFFITS when the EFFECT= or SIZE= modifier of the INFLUENCE option is not in effect. In general, an external estimate of the estimated standard error is used. When ITER > 0, the estimate is
![\[ \mr{ese}(\widehat{y}_ i) = \sqrt { \mb{x}_ i’(\mb{X}’ \mb{V}(\widehat{\btheta }_{(u)})^{-}\mb{X})^{-1}\mb{x}_ i } \]](images/statug_mixed0499.png)
When ITER=
0 and 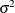 is profiled, then
is profiled, then
![\[ \mr{ese}(\widehat{y}_ i) = \widehat{\sigma }_{(u)} \sqrt { \mb{x}_ i’(\mb{X}’ \mb{V}(\widehat{\btheta }^*)^{-1}\mb{X})^{-}\mb{x}_ i} \]](images/statug_mixed0500.png)
When the EFFECT=
, SIZE=
, or KEEP=
modifier is specified, the MIXED procedure computes a multivariate version suitable for the deletion of multiple data points.
The statistic, termed MDFFITS after the MDFFIT statistic of Belsley, Kuh, and Welsch (1980, p. 32), is closely related to Cook’s D. Consider the case 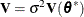 so that
so that
![\[ \mr{Var}[\widehat{\bbeta }] = \sigma ^2 (\mb{X}’ \mb{V}(\btheta ^*)^{-1}\mb{X})^{-} \]](images/statug_mixed0502.png)
and let ![$\widetilde{\mr{Var}}[\widehat{\bbeta }_{(U)}]$](images/statug_mixed0503.png) be an estimate of
be an estimate of ![$\mr{Var}[\widehat{\bbeta }_{(U)}]$](images/statug_mixed0504.png) that does not use the observations in U. The MDFFITS statistic is then computed as
that does not use the observations in U. The MDFFITS statistic is then computed as
![\[ \mr{MDFFITS}(\bbeta ) = \bdelta _{(U)}’\widetilde{\mr{Var}}[\widehat{\bbeta }_{(U)}]^{-} \bdelta _{(U)} / \mr{rank}(\mb{X}) \]](images/statug_mixed0505.png)
If ITER=
0 and 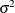 is profiled, then
is profiled, then ![$\widetilde{\mr{Var}}[\widehat{\bbeta }_{(U)}]^{-}$](images/statug_mixed0506.png) is obtained by sweeping
is obtained by sweeping
![\[ \widehat{\sigma }^2_{(U)} (\mb{X}_{(U)}’\mb{V}_{(U)}(\widehat{\btheta }^*)^{-}\mb{X}_{(U)})^{-} \]](images/statug_mixed0507.png)
The underlying idea is that if 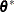 were known, then
were known, then
![\[ (\mb{X}_{(U)}’{\mb{V}_{(U)}(\btheta ^*)}^{-1}\mb{X}_{(U)})^{-} \]](images/statug_mixed0508.png)
would be ![$\mr{Var}[\widehat{\bbeta }]/\sigma ^2$](images/statug_mixed0509.png) in a generalized least squares regression with all but the data in U.
in a generalized least squares regression with all but the data in U.
In the case of iterative influence analysis, ![$\widetilde{\mr{Var}}[\widehat{\bbeta }_{(U)}]$](images/statug_mixed0503.png) is evaluated at
is evaluated at 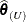 . Furthermore, a MDFFITS-type statistic is then computed for the covariance parameters:
. Furthermore, a MDFFITS-type statistic is then computed for the covariance parameters:
![\[ \mr{MDFFITS}(\btheta ) = (\widehat{\btheta } - \widehat{\btheta }_{(U)})’ \widehat{\mr{Var}}[\widehat{\btheta }_{(U)}]^{-1} (\widehat{\btheta } - \widehat{\btheta }_{(U)}) \]](images/statug_mixed0511.png)
Covariance Ratio and Trace
These statistics depend on the availability of an external estimate of 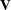 , or at least of
, or at least of 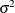 . Whereas Cook’s D and MDFFITS measure the impact of data points on a vector of parameter estimates, the covariance-based statistics measure
impact on their precision. Following Christensen, Pearson, and Johnson (1992), the MIXED procedure computes
. Whereas Cook’s D and MDFFITS measure the impact of data points on a vector of parameter estimates, the covariance-based statistics measure
impact on their precision. Following Christensen, Pearson, and Johnson (1992), the MIXED procedure computes
![\begin{align*} \mr{CovTrace}(\bbeta ) & = | \textup{trace} ( \widehat{\mr{Var}}[\widehat{\bbeta }]^{-} \, \, \widetilde{\mr{Var}}[\widehat{\bbeta }_{(U)}]) - \mr{rank}(\mb{X}) | \\ \mr{CovRatio}(\bbeta ) & = \frac{\mr{det}_{ns} (\widetilde{\mr{Var}}[\widehat{\bbeta }_{(U)}]) }{\mr{det}_{ns} (\widehat{\mr{Var}}[\widehat{\bbeta }]) } \end{align*}](images/statug_mixed0512.png)
where 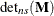 denotes the determinant of the nonsingular part of matrix
denotes the determinant of the nonsingular part of matrix 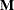 .
.
In the case of iterative influence analysis these statistics are also computed for the covariance parameter estimates. If
q denotes the rank of ![$\mr{Var}[\widehat{\btheta }]$](images/statug_mixed0515.png) , then
, then
![\begin{align*} \mr{CovTrace}(\btheta ) & = | \textup{trace}( \widehat{\mr{Var}}[\widehat{\btheta }]^{-} \, \, \widehat{\mr{Var}}[\widehat{\btheta }_{(U)}]) - q | \\ \mr{CovRatio}(\btheta ) & = \frac{\mr{det}_{ns} (\widehat{\mr{Var}}[\widehat{\btheta }_{(U)}]) }{\mr{det}_{ns} (\widehat{\mr{Var}}[\widehat{\btheta }]) } \end{align*}](images/statug_mixed0516.png)
Likelihood Distances
The log-likelihood function l and restricted log-likelihood function 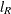 of the linear mixed model are given in the section Estimating Covariance Parameters in the Mixed Model. Denote as
of the linear mixed model are given in the section Estimating Covariance Parameters in the Mixed Model. Denote as 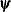 the collection of all parameters, i.e., the fixed effects
the collection of all parameters, i.e., the fixed effects 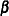 and the covariance parameters
and the covariance parameters 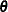 . Twice the difference between the (restricted) log-likelihood evaluated at the full-data estimates
. Twice the difference between the (restricted) log-likelihood evaluated at the full-data estimates 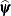 and at the reduced-data estimates
and at the reduced-data estimates 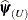 is known as the (restricted) likelihood distance:
is known as the (restricted) likelihood distance:
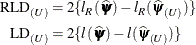
Cook and Weisberg (1982, Ch. 5.2) refer to these differences as likelihood distances, Beckman, Nachtsheim, and Cook (1987) call the measures likelihood displacements. If the number of elements in 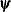 that are subject to updating following point removal is q, then likelihood displacements can be compared against cutoffs from a chi-square distribution with q degrees of freedom. Notice that this reference distribution does not depend on the number of observations removed from the
analysis, but rather on the number of model parameters that are updated. The likelihood displacement gives twice the amount
by which the log likelihood of the full data changes if one were to use an estimate based on fewer data points. It is thus
a global, summary measure of the influence of the observations in U jointly on all parameters.
that are subject to updating following point removal is q, then likelihood displacements can be compared against cutoffs from a chi-square distribution with q degrees of freedom. Notice that this reference distribution does not depend on the number of observations removed from the
analysis, but rather on the number of model parameters that are updated. The likelihood displacement gives twice the amount
by which the log likelihood of the full data changes if one were to use an estimate based on fewer data points. It is thus
a global, summary measure of the influence of the observations in U jointly on all parameters.
Unless METHOD= ML, the MIXED procedure computes the likelihood displacement based on the residual (=restricted) log likelihood, even if METHOD= MIVQUE0 or METHOD= TYPE1, TYPE2, or TYPE3.
Noniterative Update Formulas
Update formulas that do not require refitting of the model are available for the cases where 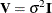 ,
, 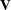 is known, or
is known, or 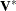 is known. When ITER=
0 and these update formulas can be invoked, the MIXED procedure uses the computational devices that are outlined in the following
paragraphs. It is then assumed that the variance-covariance matrix of the fixed effects has the form
is known. When ITER=
0 and these update formulas can be invoked, the MIXED procedure uses the computational devices that are outlined in the following
paragraphs. It is then assumed that the variance-covariance matrix of the fixed effects has the form 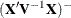 . When DDFM=
KENWARDROGER or DDFM=
KENWARDROGER2, this is not the case; the estimated variance-covariance matrix is then inflated to better represent the uncertainty
in the estimated covariance parameters. Influence statistics when DDFM=
KENWARDROGER should iteratively update the covariance parameters (ITER
> 0). The dependence of
. When DDFM=
KENWARDROGER or DDFM=
KENWARDROGER2, this is not the case; the estimated variance-covariance matrix is then inflated to better represent the uncertainty
in the estimated covariance parameters. Influence statistics when DDFM=
KENWARDROGER should iteratively update the covariance parameters (ITER
> 0). The dependence of 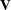 on
on 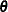 is suppressed in the sequel for brevity.
is suppressed in the sequel for brevity.
Updating the Fixed Effects
Denote by 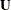 the
the 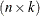 matrix that is assembled from k columns of the identity matrix. Each column of
matrix that is assembled from k columns of the identity matrix. Each column of 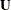 corresponds to the removal of one data point. The point being targeted by the ith column of
corresponds to the removal of one data point. The point being targeted by the ith column of 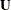 corresponds to the row in which a 1 appears. Furthermore, define
corresponds to the row in which a 1 appears. Furthermore, define
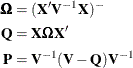
The change in the fixed-effects estimates following removal of the observations in U is
![\[ \widehat{\bbeta } - \widehat{\bbeta }_{(U)} = \bOmega \mb{X}’\mb{V}^{-1}\mb{U} (\mb{U}’\mb{P}\mb{U})^{-1}\mb{U}’\mb{V}^{-1} (\mb{y}-\mb{X}\widehat{\bbeta }) \]](images/statug_mixed0528.png)
Using results in Cook and Weisberg (1982, A2) you can further compute
![\[ \widetilde{\bOmega } = (\mb{X}_{(U)}’\mb{V}_{(U)}^{-1}\mb{X}_{(U)})^{-} = \bOmega + \bOmega \mb{X}’\mb{V}^{-1}\mb{U} (\mb{U}’\mb{P}\mb{U})^{-1} \mb{U}’\mb{V}^{-1}\mb{X}\bOmega \]](images/statug_mixed0529.png)
If 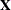 is
is 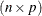 of rank
of rank 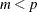 , then
, then 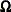 is deficient in rank and the MIXED procedure computes needed quantities in
is deficient in rank and the MIXED procedure computes needed quantities in 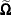 by sweeping (Goodnight 1979). If the rank of the
by sweeping (Goodnight 1979). If the rank of the 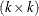 matrix
matrix 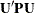 is less than k, the removal of the observations introduces a new singularity, whether
is less than k, the removal of the observations introduces a new singularity, whether 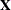 is of full rank or not. The solution vectors
is of full rank or not. The solution vectors 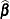 and
and 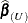 then do not have the same expected values and should not be compared. When the MIXED procedure encounters this situation,
influence diagnostics that depend on the choice of generalized inverse are not computed. The procedure also monitors the singularity
criteria when sweeping the rows of
then do not have the same expected values and should not be compared. When the MIXED procedure encounters this situation,
influence diagnostics that depend on the choice of generalized inverse are not computed. The procedure also monitors the singularity
criteria when sweeping the rows of 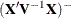 and of
and of 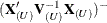 . If a new singularity is encountered or a former singularity disappears, no influence statistics are computed.
. If a new singularity is encountered or a former singularity disappears, no influence statistics are computed.
Residual Variance
When 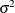 is profiled out of the marginal variance-covariance matrix, a closed-form estimate of
is profiled out of the marginal variance-covariance matrix, a closed-form estimate of 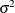 that is based on only the remaining observations can be computed provided
that is based on only the remaining observations can be computed provided 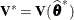 is known. Hurtado (1993, Thm. 5.2) shows that
is known. Hurtado (1993, Thm. 5.2) shows that
![\[ (n-q-r)\widehat{\sigma }^2_{(U)} = (n-q)\widehat{\sigma }^2 - \widehat{\bepsilon }_ U’(\widehat{\sigma }^2 \mb{U}’\bP \mb{U})^{-1}\widehat{\bepsilon }_ U \]](images/statug_mixed0539.png)
and 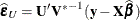 . In the case of maximum likelihood estimation q = 0 and for REML estimation
. In the case of maximum likelihood estimation q = 0 and for REML estimation 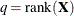 . The constant r equals the rank of
. The constant r equals the rank of 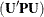 for REML estimation and the number of effective observations that are removed if METHOD=
ML.
for REML estimation and the number of effective observations that are removed if METHOD=
ML.
Likelihood Distances
For noniterative methods the following computational devices are used to compute (restricted) likelihood distances provided
that the residual variance 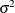 is profiled.
is profiled.
The log likelihood function 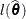 evaluated at the full-data and reduced-data estimates can be written as
evaluated at the full-data and reduced-data estimates can be written as
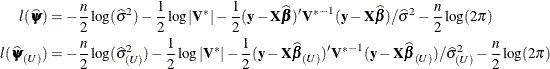
Notice that 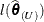 evaluates the log likelihood for n data points at the reduced-data estimates. It is not the log likelihood obtained by fitting the model to the reduced data.
The likelihood distance is then
evaluates the log likelihood for n data points at the reduced-data estimates. It is not the log likelihood obtained by fitting the model to the reduced data.
The likelihood distance is then
![\[ \mr{LD}_{(U)} = n\log \left\{ \frac{\widehat{\sigma }^2_{(U)}}{\widehat{\sigma }^2} \right\} - n + \left(\mb{y}-\mb{X}\widehat{\bbeta }_{(U)}\right)’{\mb{V}^*}^{-1} \left(\mb{y}-\mb{X}\widehat{\bbeta }_{(U)}\right) / \widehat{\sigma }^2_{(U)} \]](images/statug_mixed0546.png)
Expressions for 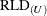 in noniterative influence analysis are derived along the same lines.
in noniterative influence analysis are derived along the same lines.

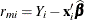
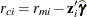
![$r_{mi}^{\mathit{student}} = \frac{r_{mi}}{\sqrt {\widehat{\mr{Var}}[r_{mi}]}}$](images/statug_mixed0429.png)
![$r_{ci}^{\mathit{student}} = \frac{r_{ci}}{\sqrt {\widehat{\mr{Var}}[r_{ci}]}}$](images/statug_mixed0430.png)
![$r_{mi}^{\mathit{pearson}} = \frac{r_{mi}}{\sqrt {\widehat{\mr{Var}}[Y_ i]}}$](images/statug_mixed0431.png)
![$r_{ci}^{\mathit{pearson}} = \frac{r_{ci}}{\sqrt {\widehat{\mr{Var}}[Y_ i|\bgamma ]}}$](images/statug_mixed0432.png)