The MIANALYZE Procedure
- Overview
- Getting Started
-
Syntax

-
Details

-
Examples
 Reading Means and Standard Errors from a DATA= Data SetReading Means and Covariance Matrices from a DATA= COV Data SetReading Regression Results from a DATA= EST Data SetReading Mixed Model Results from PARMS= and COVB= Data SetsReading Generalized Linear Model ResultsReading GLM Results from PARMS= and XPXI= Data SetsReading Logistic Model Results from a PARMS= Data SetReading Mixed Model Results with Classification CovariatesReading Nominal Logistic Model ResultsUsing a TEST statementCombining Correlation CoefficientsSensitivity Analysis with Control-Based Pattern ImputationSensitivity Analysis with the Tipping-Point Approach
Reading Means and Standard Errors from a DATA= Data SetReading Means and Covariance Matrices from a DATA= COV Data SetReading Regression Results from a DATA= EST Data SetReading Mixed Model Results from PARMS= and COVB= Data SetsReading Generalized Linear Model ResultsReading GLM Results from PARMS= and XPXI= Data SetsReading Logistic Model Results from a PARMS= Data SetReading Mixed Model Results with Classification CovariatesReading Nominal Logistic Model ResultsUsing a TEST statementCombining Correlation CoefficientsSensitivity Analysis with Control-Based Pattern ImputationSensitivity Analysis with the Tipping-Point Approach - References
Example 76.6 Reading GLM Results from PARMS= and XPXI= Data Sets
This example creates data sets that contains parameter estimates and corresponding 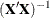 matrices computed by a general linear model analysis for a set of imputed data sets. These estimates are then combined to
generate valid statistical inferences about the model parameters.
matrices computed by a general linear model analysis for a set of imputed data sets. These estimates are then combined to
generate valid statistical inferences about the model parameters.
The following statements use PROC GLM to generate the parameter estimates and 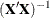 matrix for each imputed data set:
matrix for each imputed data set:
ods select none;
proc glm data=outmi;
model Oxygen= RunTime RunPulse/inverse;
by _Imputation_;
ods output ParameterEstimates=glmparms
InvXPX=glmxpxi;
quit;
ods select all;
Because of the ODS SELECT statements, no output is displayed. The following statements display (in Output 76.6.1) the output parameter estimates and standard errors from PROC GLM for the first two imputed data sets:
proc print data=glmparms (obs=6); var _Imputation_ Parameter Estimate StdErr; title 'GLM Model Coefficients (First Two Imputations)'; run;
Output 76.6.1: PROC GLM Model Coefficients
| GLM Model Coefficients (First Two Imputations) |
| Obs | _Imputation_ | Parameter | Estimate | StdErr |
|---|---|---|---|---|
| 1 | 1 | Intercept | 86.5440339 | 10.00726811 |
| 2 | 1 | RunTime | -2.8223108 | 0.32824165 |
| 3 | 1 | RunPulse | -0.0587292 | 0.05854109 |
| 4 | 2 | Intercept | 83.0207303 | 8.88996885 |
| 5 | 2 | RunTime | -3.0002288 | 0.33847204 |
| 6 | 2 | RunPulse | -0.0249103 | 0.05137859 |
The following statements display (in Output 76.6.2) 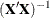 matrices from PROC GLM for the first two imputed data sets:
matrices from PROC GLM for the first two imputed data sets:
proc print data=glmxpxi (obs=8); var _Imputation_ Parameter Intercept RunTime RunPulse; title 'GLM X''X Inverse Matrices (First Two Imputations)'; run;
Output 76.6.2: PROC GLM 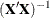 Matrices
Matrices
| GLM X'X Inverse Matrices (First Two Imputations) |
| Obs | _Imputation_ | Parameter | Intercept | RunTime | RunPulse |
|---|---|---|---|---|---|
| 1 | 1 | Intercept | 12.696250656 | -0.067849956 | -0.069826009 |
| 2 | 1 | RunTime | -0.067849956 | 0.0136594055 | -0.000436938 |
| 3 | 1 | RunPulse | -0.069826009 | -0.000436938 | 0.0004344762 |
| 4 | 1 | Oxygen | 86.544033929 | -2.822310769 | -0.058729234 |
| 5 | 2 | Intercept | 10.784620785 | -0.091107072 | -0.057201387 |
| 6 | 2 | RunTime | -0.091107072 | 0.0156332765 | -0.000426902 |
| 7 | 2 | RunPulse | -0.057201387 | -0.000426902 | 0.0003602208 |
| 8 | 2 | Oxygen | 83.020730343 | -3.000228818 | -0.024910305 |
The standard errors for the estimates in the output Glmparms data set are needed to create the covariance matrix from the 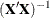 matrix. The following statements use the MIANALYZE procedure with input PARMS= and XPXI= data sets to produce the same results
as displayed in Output 76.3.2 and Output 76.3.3 in Example 76.3:
matrix. The following statements use the MIANALYZE procedure with input PARMS= and XPXI= data sets to produce the same results
as displayed in Output 76.3.2 and Output 76.3.3 in Example 76.3:
proc mianalyze parms=glmparms xpxi=glmxpxi edf=28; modeleffects Intercept RunTime RunPulse; run;