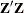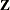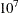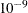The ICPHREG Procedure
PROC ICPHREG Statement
-
PROC ICPHREG <options>;
The PROC ICPHREG statement invokes the ICPHREG procedure. Table 63.2 summarizes the options available in the PROC ICPHREG statement.
Table 63.2: PROC ICPHREG Statement Options
|
Option |
Description |
|---|---|
|
Specifies the level for confidence limits |
|
|
Names the SAS data set to be analyzed |
|
|
Displays the iteration history, final gradient, and second derivative matrix |
|
|
Specifies the length of effect names |
|
|
Specifies optimization parameters for fitting the specified model |
|
|
Suppresses all displayed output |
|
|
Requests a single-threaded mode for the computation |
|
|
Controls the plots that are produced through ODS Graphics |
|
|
Specifies the singularity tolerance |
|
|
Specifies the number of threads for the computation |
You can specify the following options in the PROC ICPHREG statement.


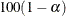
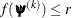
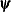
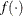
![\[ |f(\bpsi ^{(k-1)}) - f(\bpsi ^{(k)})| \leq r \]](images/statug_icphreg0012.png)
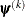
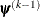
![\[ \max _ j |g_ j(\bpsi ^{(k)})| \leq r \]](images/statug_icphreg0015.png)
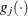
![\[ \frac{|f(\bpsi ^{(k)}) - f(\bpsi ^{(k-1)})|}{|f(\bpsi ^{(k-1)})|} \leq r \]](images/statug_icphreg0017.png)
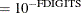
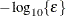

![\[ \frac{\mb{g}(\bpsi ^{(k)})^\prime [\bH ^{(k)}]^{-1} \mb{g}(\bpsi ^{(k)})}{|f(\bpsi ^{(k)})| } \leq r \]](images/statug_icphreg0021.png)
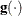
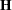
![\[ \frac{\parallel \mb{g}(\bpsi ^{(k)}) \parallel _2^2 \quad \parallel \mb{g}(\bpsi ^{(k)}) \parallel _2}{\parallel \mb{g}(\bpsi ^{(k)}) - \mb{g}(\bpsi ^{(k-1)}) \parallel _2 |f(\bpsi ^{(k)})| } \leq r \]](images/statug_icphreg0024.png)