The ICPHREG Procedure
Getting Started: ICPHREG Procedure
This example demonstrates how you can fit a proportional hazards model on an interval-censored data set. By default, PROC ICPHREG uses a piecewise constant baseline hazard to fit the model.
The AIDS data (Larder, Darby, and Richman 1989) consist of observations from 31 patients who were followed up for the development of drug resistance to zidovudine. The
following DATA step creates the SAS data set HIV:
data hiv; input Left Right Stage Dose CdLow CdHigh; if (Left=0) then Left=.; if (Right>=26) then Right=.; datalines; 0 16 0 0 0 1 15 26 0 0 0 1 12 26 0 0 0 1 17 26 0 0 0 1 13 26 0 0 0 1 0 24 0 0 1 0 6 26 0 1 1 0 0 15 0 1 1 0 14 26 0 1 1 0 12 26 0 1 1 0 13 26 0 1 0 1 12 26 0 1 1 0 12 26 0 1 1 0 0 18 0 1 0 1 0 14 0 1 0 1 0 17 0 1 1 0 0 15 0 1 1 0 3 26 1 0 0 1 4 26 1 0 0 1 1 11 1 0 0 1 13 19 1 0 0 1 0 6 1 0 0 1 0 11 1 1 0 0 6 26 1 1 0 0 0 6 1 1 0 0 2 12 1 1 0 0 1 17 1 1 1 0 0 14 1 1 0 0 0 25 1 1 0 1 2 11 1 1 0 0 0 14 1 1 0 0 ;
The data set HIV contains the variables Left and Right, which are the starting time and ending time, both in months since the start of study; the variable Stage, which indicates the stage of disease (early (0) or late (1)); the variable Dose, a binary variable that indicates whether the dose is low (0) or high (1); the variable CdLow, which indicates whether the CD4 lymphocyte count is less than 100; and the variable CdHigh, which indicates that a count greater than or equal to 400 is recorded.
The following statements use PROC ICPHREG to fit a proportional hazards model to these data:
proc icphreg data=hiv; class Stage Dose / desc; model (Left, Right) = Stage Dose; run;
The CLASS statement specifies that the variables Stage and Dose are classification variables. The DESC option sets the lower formatted value as the reference level for each CLASS variable.
The MODEL statement specifies that the observed intervals are formed by Left and Right.
By default, the preceding statements produce information about the input data and the fitted model, as shown in Figure 63.1.
Figure 63.1: Model and Data Information from the ICPHREG Procedure
Figure 63.1 shows 13 left-censored observations, 13 right-censored observations, and 5 interval-censored observations.
Figure 63.2 displays the "Class Level Information" table, which identifies the levels of the classification variables that are used in the model.
Figure 63.2: CLASS Variables Information from the ICPHREG Procedure
By default, PROC ICPHREG uses a baseline hazard that is partitioned into five disjoint intervals to fit a proportional hazards model. Figure 63.3 displays details about this partition.
Figure 63.3: Interval Partition
PROC ICPHREG determines the break points so that each time interval contains approximately an equal number of imputed middle points and boundary values in the input data set after excluding the right-censored observations. For more information about this method, see the section Choosing Break Points. You can supply your own partition by using the INTERVALS= option in the MODEL statement.
The "Fit Statistics" table, shown in Figure 63.4, contains several statistics that summarize how well the model fits the data. These statistics are helpful in judging the adequacy of a model and in comparing it with other models under consideration.
Figure 63.4: Model Fit Statistics from the ICPHREG Procedure
The table of parameter estimates is displayed in Figure 63.5. The columns display the parameter name, the degrees of freedom that are associated with the parameter, the estimated parameter value, the standard error of the parameter estimate, the confidence limits, the Wald chi-square statistic, and the associated p-value for testing the significance of the parameter. If a parameter has been fixed during the optimization process, or if a column of the Hessian matrix that corresponds to that parameter is found to linearly depend on columns that correspond to proceeding model parameters, PROC ICPHREG assigns zero degrees of freedom to that parameter and displays a value of zero for its standard error.
Figure 63.5: Model Parameter Estimates from the ICPHREG Procedure
| Analysis of Maximum Likelihood Parameter Estimates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect | Stage | Dose | DF | Estimate | Standard Error |
95% Confidence Limits | Chi-Square | Pr > ChiSq | |
| Haz1 | 0 | 0.0000 | |||||||
| Haz2 | 1 | 0.0167 | 0.0205 | 0.0000 | 0.0568 | ||||
| Haz3 | 0 | 0.0000 | |||||||
| Haz4 | 1 | 0.0842 | 0.0655 | 0.0000 | 0.2126 | ||||
| Haz5 | 1 | 2.5641 | 366.4263 | 0.0000 | 720.7464 | ||||
| Stage | 1 | 1 | 2.9597 | 0.9358 | 1.1255 | 4.7939 | 10.00 | 0.0016 | |
| Stage | 0 | 0 | 0.0000 | ||||||
| Dose | 1 | 1 | 1.6229 | 0.8410 | -0.0255 | 3.2713 | 3.72 | 0.0537 | |
| Dose | 0 | 0 | 0.0000 | ||||||
Two types of parameters are present in Figure 63.5: the hazard parameters (Haz1, Haz2, ..., Haz5) and the regression coefficients for the covariates. PROC ICPHREG does not display the chi-square statistic and associated p-value for the hazard parameters.
Two of the hazard parameters are constrained at 0, a sign of overparameterization that results from too many hazard parameters in the model. For more information about how the constraints are constructed, see the section NOPOLISH. You can use fewer break points to fit the model by using the NINTERVAL= option or the INTERVALS= option. For example, the following statements request a model that has exactly two hazard parameters by specifying one break point at 10:
proc icphreg data=hiv ithistory; class Stage Dose / desc; model (Left, Right) = Stage Dose / basehaz=pch(intervals=(10)); run;
The table of parameter estimates is displayed in Figure 63.6. None of the hazard parameters are constrained.
Figure 63.6: Model Parameter Estimates from the ICPHREG Procedure
| Analysis of Maximum Likelihood Parameter Estimates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect | Stage | Dose | DF | Estimate | Standard Error |
95% Confidence Limits | Chi-Square | Pr > ChiSq | |
| Haz1 | 1 | 0.0042 | 0.0051 | 0.0000 | 0.0142 | ||||
| Haz2 | 1 | 0.0590 | 0.0360 | 0.0000 | 0.1296 | ||||
| Stage | 1 | 1 | 2.0810 | 0.7298 | 0.6506 | 3.5114 | 8.13 | 0.0044 | |
| Stage | 0 | 0 | 0.0000 | ||||||
| Dose | 1 | 1 | 1.0907 | 0.6766 | -0.2354 | 2.4167 | 2.60 | 0.1069 | |
| Dose | 0 | 0 | 0.0000 | ||||||
The ITHISTORY option outputs the iteration history of the fitting algorithm, which is shown in Figure 63.7. This option also produces the gradient and Hessian of the likelihood function at the last evaluation. In Figure 63.7, all values of the gradient are close to zero.
Figure 63.7: Iteration History from the ICPHREG Procedure
| Likelihood Optimization Iteration History | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Iteration | Evaluations | -2 Log Likelihood |
Change | Max Gradient |
Parameter Values | Gradient Values | ||||||
| Stage1 | Dose1 | Haz1 | Haz2 | Stage1 | Dose1 | Haz1 | Haz2 | |||||
| 0 | 2 | 47.8895 | . | 74.9948 | 0 | 0 | 0.1245 | 0.0741 | -1.8827 | 3.6394 | 74.9948 | 5.0972 |
| 1 | 10 | 39.1668 | -8.7227 | 47.5072 | 0.1816 | 0.3012 | 0.0559 | 0.0848 | -3.6580 | 0.5057 | 47.5072 | -1.3723 |
| 2 | 6 | 37.6893 | -1.4775 | 32.3796 | 0.2453 | 0.3347 | 0.0440 | 0.0944 | -3.9790 | -0.2883 | 32.3796 | -1.7277 |
| 3 | 3 | 35.3576 | -2.3317 | 50.9346 | 0.4701 | 0.4206 | 0.0185 | 0.1168 | -4.6140 | -2.0073 | -50.9346 | -3.9286 |
| 4 | 3 | 32.5990 | -2.7586 | 6.4037 | 0.8039 | 0.5075 | 0.0200 | 0.1003 | -2.8566 | -0.7001 | 6.4037 | 0.3183 |
| 5 | 3 | 30.1026 | -2.4964 | 94.9226 | 1.3800 | 0.7032 | 0.00785 | 0.0839 | -2.1446 | -1.3702 | -94.9226 | -6.0631 |
| 6 | 3 | 29.0224 | -1.0802 | 23.8304 | 1.7623 | 0.8588 | 0.00724 | 0.0699 | -0.3881 | -0.2204 | 23.8304 | -0.6369 |
| 7 | 3 | 28.8561 | -0.1663 | 103.7 | 2.0115 | 1.0376 | 0.00384 | 0.0622 | -0.5201 | -0.4868 | -103.7 | -2.6791 |
| 8 | 3 | 28.7697 | -0.0863 | 1.9794 | 2.0740 | 1.0858 | 0.00418 | 0.0593 | -0.0194 | -0.0145 | -1.9794 | -0.0585 |
| 9 | 3 | 28.7696 | -0.00013 | 0.00309 | 2.0799 | 1.0898 | 0.00416 | 0.0590 | -0.00170 | -0.00121 | 0.00309 | -0.00077 |
| 10 | 3 | 28.7696 | -3.01E-6 | 0.000803 | 2.0809 | 1.0906 | 0.00416 | 0.0590 | -0.00015 | -0.00012 | -0.00080 | -0.00004 |
| 11 | 3 | 28.7696 | -2.5E-8 | 6.991E-6 | 2.0810 | 1.0907 | 0.00416 | 0.0590 | -6.99E-6 | -5.67E-6 | -6.93E-6 | -1.38E-7 |
| 12 | 2 | 28.7696 | 0 | 6.991E-6 | 2.0810 | 1.0907 | 0.00416 | 0.0590 | -6.99E-6 | -5.67E-6 | -6.93E-6 | -1.38E-7 |
One reason for fitting a proportional hazards model is to evaluate the hazard ratios between various disease groups. You can request customized hazard ratios by using the HAZARDRATIO statement, as follows:
proc icphreg data=hiv; class Stage / desc; model (Left, Right) = Stage / basehaz=pch(intervals=(10)); hazardratio Stage; run;
Figure 63.8 shows the estimated hazard ratio between the values 1 and 0 of the Stage variable and the corresponding 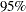 confidence limits.
confidence limits.
Figure 63.8: Hazard Ratio Estimate between Stage Values 1 and 0
The estimate of 5.624 indicates that patients who have Stage 1 disease tend to have a much higher risk of developing AIDS than those who have Stage 0. However, the confidence limits are wide because of the small sample size.
