The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD Statement ASSESS Statement BAYES Statement BY Statement CLASS Statement CONTRAST Statement DEVIANCE Statement EFFECTPLOT Statement ESTIMATE Statement EXACT Statement EXACTOPTIONS Statement FREQ Statement FWDLINK Statement INVLINK Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement OUTPUT Statement Programming Statements REPEATED Statement SLICE Statement STORE Statement STRATA Statement VARIANCE Statement WEIGHT Statement ZEROMODEL Statement
PROC GENMOD Statement ASSESS Statement BAYES Statement BY Statement CLASS Statement CONTRAST Statement DEVIANCE Statement EFFECTPLOT Statement ESTIMATE Statement EXACT Statement EXACTOPTIONS Statement FREQ Statement FWDLINK Statement INVLINK Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement OUTPUT Statement Programming Statements REPEATED Statement SLICE Statement STORE Statement STRATA Statement VARIANCE Statement WEIGHT Statement ZEROMODEL Statement -
Details
 Generalized Linear Models Theory Specification of Effects Parameterization Used in PROC GENMOD Type 1 Analysis Type 3 Analysis Confidence Intervals for Parameters F Statistics Lagrange Multiplier Statistics Predicted Values of the Mean Residuals Multinomial Models Zero-Inflated Models Generalized Estimating Equations Assessment of Models Based on Aggregates of Residuals Case Deletion Diagnostic Statistics Bayesian Analysis Exact Logistic and Exact Poisson Regression Missing Values Displayed Output for Classical Analysis Displayed Output for Bayesian Analysis Displayed Output for Exact Analysis ODS Table Names ODS Graphics
Generalized Linear Models Theory Specification of Effects Parameterization Used in PROC GENMOD Type 1 Analysis Type 3 Analysis Confidence Intervals for Parameters F Statistics Lagrange Multiplier Statistics Predicted Values of the Mean Residuals Multinomial Models Zero-Inflated Models Generalized Estimating Equations Assessment of Models Based on Aggregates of Residuals Case Deletion Diagnostic Statistics Bayesian Analysis Exact Logistic and Exact Poisson Regression Missing Values Displayed Output for Classical Analysis Displayed Output for Bayesian Analysis Displayed Output for Exact Analysis ODS Table Names ODS Graphics -
Examples
 Logistic Regression Normal Regression, Log Link Gamma Distribution Applied to Life Data Ordinal Model for Multinomial Data GEE for Binary Data with Logit Link Function Log Odds Ratios and the ALR Algorithm Log-Linear Model for Count Data Model Assessment of Multiple Regression Using Aggregates of Residuals Assessment of a Marginal Model for Dependent Data Bayesian Analysis of a Poisson Regression Model Exact Poisson Regression
Logistic Regression Normal Regression, Log Link Gamma Distribution Applied to Life Data Ordinal Model for Multinomial Data GEE for Binary Data with Logit Link Function Log Odds Ratios and the ALR Algorithm Log-Linear Model for Count Data Model Assessment of Multiple Regression Using Aggregates of Residuals Assessment of a Marginal Model for Dependent Data Bayesian Analysis of a Poisson Regression Model Exact Poisson Regression - References
Example 39.2 Normal Regression, Log Link
Consider the following data, where x is an explanatory variable and y is the response variable. It appears that y varies nonlinearly with x and that the variance is approximately constant. A normal distribution with a log link function is chosen to model these data; that is, 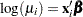 so that
so that 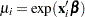 .
.
data nor; input x y; datalines; 0 5 0 7 0 9 1 7 1 10 1 8 2 11 2 9 3 16 3 13 3 14 4 25 4 24 5 34 5 32 5 30 ;
The following SAS statements produce the analysis with the normal distribution and log link:
proc genmod data=nor;
model y = x / dist = normal
link = log;
output out = Residuals
pred = Pred
resraw = Resraw
reschi = Reschi
resdev = Resdev
stdreschi = Stdreschi
stdresdev = Stdresdev
reslik = Reslik;
run;
The OUTPUT statement is specified to produce a data set that contains predicted values and residuals for each observation. This data set can be useful for further analysis, such as residual plotting.
The results from these statements are displayed in Output 39.2.1.
| Model Information | |
|---|---|
| Data Set | WORK.NOR |
| Distribution | Normal |
| Link Function | Log |
| Dependent Variable | y |
| Criteria For Assessing Goodness Of Fit | |||
|---|---|---|---|
| Criterion | DF | Value | Value/DF |
| Deviance | 14 | 52.3000 | 3.7357 |
| Scaled Deviance | 14 | 16.0000 | 1.1429 |
| Pearson Chi-Square | 14 | 52.3000 | 3.7357 |
| Scaled Pearson X2 | 14 | 16.0000 | 1.1429 |
| Log Likelihood | -32.1783 | ||
| Full Log Likelihood | -32.1783 | ||
| AIC (smaller is better) | 70.3566 | ||
| AICC (smaller is better) | 72.3566 | ||
| BIC (smaller is better) | 72.6743 | ||
| Analysis Of Maximum Likelihood Parameter Estimates | |||||||
|---|---|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error | Wald 95% Confidence Limits | Wald Chi-Square | Pr > ChiSq | |
| Intercept | 1 | 1.7214 | 0.0894 | 1.5461 | 1.8966 | 370.76 | <.0001 |
| x | 1 | 0.3496 | 0.0206 | 0.3091 | 0.3901 | 286.64 | <.0001 |
| Scale | 1 | 1.8080 | 0.3196 | 1.2786 | 2.5566 | ||
| Note: | The scale parameter was estimated by maximum likelihood. |
The PROC GENMOD scale parameter, in the case of the normal distribution, is the standard deviation. By default, the scale parameter is estimated by maximum likelihood. You can specify a fixed standard deviation by using the NOSCALE and SCALE= options in the MODEL statement.
proc print data=Residuals ; run;
| Obs | x | y | Pred | Reschi | Resraw | Resdev | Stdreschi | Stdresdev | Reslik |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 5 | 5.5921 | -0.59212 | -0.59212 | -0.59212 | -0.34036 | -0.34036 | -0.34036 |
| 2 | 0 | 7 | 5.5921 | 1.40788 | 1.40788 | 1.40788 | 0.80928 | 0.80928 | 0.80928 |
| 3 | 0 | 9 | 5.5921 | 3.40788 | 3.40788 | 3.40788 | 1.95892 | 1.95892 | 1.95892 |
| 4 | 1 | 7 | 7.9324 | -0.93243 | -0.93243 | -0.93243 | -0.54093 | -0.54093 | -0.54093 |
| 5 | 1 | 10 | 7.9324 | 2.06757 | 2.06757 | 2.06757 | 1.19947 | 1.19947 | 1.19947 |
| 6 | 1 | 8 | 7.9324 | 0.06757 | 0.06757 | 0.06757 | 0.03920 | 0.03920 | 0.03920 |
| 7 | 2 | 11 | 11.2522 | -0.25217 | -0.25217 | -0.25217 | -0.14686 | -0.14686 | -0.14686 |
| 8 | 2 | 9 | 11.2522 | -2.25217 | -2.25217 | -2.25217 | -1.31166 | -1.31166 | -1.31166 |
| 9 | 3 | 16 | 15.9612 | 0.03878 | 0.03878 | 0.03878 | 0.02249 | 0.02249 | 0.02249 |
| 10 | 3 | 13 | 15.9612 | -2.96122 | -2.96122 | -2.96122 | -1.71738 | -1.71738 | -1.71738 |
| 11 | 3 | 14 | 15.9612 | -1.96122 | -1.96122 | -1.96122 | -1.13743 | -1.13743 | -1.13743 |
| 12 | 4 | 25 | 22.6410 | 2.35897 | 2.35897 | 2.35897 | 1.37252 | 1.37252 | 1.37252 |
| 13 | 4 | 24 | 22.6410 | 1.35897 | 1.35897 | 1.35897 | 0.79069 | 0.79069 | 0.79069 |
| 14 | 5 | 34 | 32.1163 | 1.88366 | 1.88366 | 1.88366 | 1.22914 | 1.22914 | 1.22914 |
| 15 | 5 | 32 | 32.1163 | -0.11634 | -0.11634 | -0.11634 | -0.07592 | -0.07592 | -0.07592 |
| 16 | 5 | 30 | 32.1163 | -2.11634 | -2.11634 | -2.11634 | -1.38098 | -1.38098 | -1.38098 |
The data set of predicted values and residuals (Output 39.2.2) is created by the OUTPUT statement. You can use the PLOTS= option in the PROC GENMOD statement to create plots of predicted values and residuals. Note that raw, Pearson, and deviance residuals are equal in this example. This is a characteristic of the normal distribution and is not true in general for other distributions.