The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD Statement ASSESS Statement BAYES Statement BY Statement CLASS Statement CONTRAST Statement DEVIANCE Statement EFFECTPLOT Statement ESTIMATE Statement EXACT Statement EXACTOPTIONS Statement FREQ Statement FWDLINK Statement INVLINK Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement OUTPUT Statement Programming Statements REPEATED Statement SLICE Statement STORE Statement STRATA Statement VARIANCE Statement WEIGHT Statement ZEROMODEL Statement
PROC GENMOD Statement ASSESS Statement BAYES Statement BY Statement CLASS Statement CONTRAST Statement DEVIANCE Statement EFFECTPLOT Statement ESTIMATE Statement EXACT Statement EXACTOPTIONS Statement FREQ Statement FWDLINK Statement INVLINK Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement OUTPUT Statement Programming Statements REPEATED Statement SLICE Statement STORE Statement STRATA Statement VARIANCE Statement WEIGHT Statement ZEROMODEL Statement -
Details
 Generalized Linear Models Theory Specification of Effects Parameterization Used in PROC GENMOD Type 1 Analysis Type 3 Analysis Confidence Intervals for Parameters F Statistics Lagrange Multiplier Statistics Predicted Values of the Mean Residuals Multinomial Models Zero-Inflated Models Generalized Estimating Equations Assessment of Models Based on Aggregates of Residuals Case Deletion Diagnostic Statistics Bayesian Analysis Exact Logistic and Exact Poisson Regression Missing Values Displayed Output for Classical Analysis Displayed Output for Bayesian Analysis Displayed Output for Exact Analysis ODS Table Names ODS Graphics
Generalized Linear Models Theory Specification of Effects Parameterization Used in PROC GENMOD Type 1 Analysis Type 3 Analysis Confidence Intervals for Parameters F Statistics Lagrange Multiplier Statistics Predicted Values of the Mean Residuals Multinomial Models Zero-Inflated Models Generalized Estimating Equations Assessment of Models Based on Aggregates of Residuals Case Deletion Diagnostic Statistics Bayesian Analysis Exact Logistic and Exact Poisson Regression Missing Values Displayed Output for Classical Analysis Displayed Output for Bayesian Analysis Displayed Output for Exact Analysis ODS Table Names ODS Graphics -
Examples
 Logistic Regression Normal Regression, Log Link Gamma Distribution Applied to Life Data Ordinal Model for Multinomial Data GEE for Binary Data with Logit Link Function Log Odds Ratios and the ALR Algorithm Log-Linear Model for Count Data Model Assessment of Multiple Regression Using Aggregates of Residuals Assessment of a Marginal Model for Dependent Data Bayesian Analysis of a Poisson Regression Model Exact Poisson Regression
Logistic Regression Normal Regression, Log Link Gamma Distribution Applied to Life Data Ordinal Model for Multinomial Data GEE for Binary Data with Logit Link Function Log Odds Ratios and the ALR Algorithm Log-Linear Model for Count Data Model Assessment of Multiple Regression Using Aggregates of Residuals Assessment of a Marginal Model for Dependent Data Bayesian Analysis of a Poisson Regression Model Exact Poisson Regression - References
| REPEATED Statement |
- REPEATED SUBJECT= subject-effect </ options> ;
The REPEATED statement specifies the covariance structure of multivariate responses for GEE model fitting in the GENMOD procedure. In addition, the REPEATED statement controls the iterative fitting algorithm used in GEEs and specifies optional output. Other GENMOD procedure statements, such as the MODEL and CLASS statements, are used in the same way as they are for ordinary generalized linear models to specify the regression model for the mean of the responses.
- SUBJECT=subject-effect
-
identifies subjects in the input data set. The subject-effect can be a single variable, an interaction effect, a nested effect, or a combination. Each distinct value, or level, of the effect identifies a different subject, or cluster. Responses from different subjects are assumed to be statistically independent, and responses within subjects are assumed to be correlated. A subject-effect must be specified, and variables used in defining the subject-effect must be listed in the CLASS statement. The input data set does not need to be sorted by subject (see the SORTED option).
The options control how the model is fit and what output is produced. You can specify the following options after a slash (/).
- ALPHAINIT=numbers
specifies initial values for log odds ratio regression parameters if the LOGOR= option is specified for binary data. If this option is not specified, an initial value of 0.01 is used for all the parameters.
- CONVERGE=number
specifies the convergence criterion for GEE parameter estimation. If the maximum absolute difference between regression parameter estimates is less than the value of number on two successive iterations, convergence is declared. If the absolute value of a regression parameter estimate is greater than 0.08, then the absolute difference normalized by the regression parameter value is used instead of the absolute difference. The default value of number is 0.0001.
- CORRW
displays the estimated working correlation matrix. If you specify an exchangeable working correlation structure with the CORR=EXCH option, the CORRW option is not needed to view the estimated correlation, since a table is printed by default that contains the single estimated correlation.
- CORRB
displays the estimated regression parameter correlation matrix. Both model-based and empirical correlations are displayed.
- COVB
displays the estimated regression parameter covariance matrix. Both model-based and empirical covariances are displayed.
- ECORRB
displays the estimated regression parameter empirical correlation matrix.
- ECOVB
displays the estimated regression parameter empirical covariance matrix.
- INTERCEPT=number
specifies either an initial or a fixed value of the intercept regression parameter in the GEE model. If you specify the NOINT option in the MODEL statement, then the intercept is fixed at the value of number.
- INITIAL=numbers
specifies initial values of the regression parameters estimation, other than the intercept parameter, for GEE estimation. If this option is not specified, the estimated regression parameters assuming independence for all responses are used for the initial values.
- LOGOR=log-odds-ratio-structure-keyword
-
specifies the regression structure of the log odds ratio used to model the association of the responses from subjects for binary data. The response syntax must be of the single variable type, the distribution must be binomial, and the data must be binary. Table 39.5 displays the log odds ratio structure keywords and the corresponding log odds ratio regression structures. See the section Alternating Logistic Regressions for definitions of the log odds ratio types and examples of specifying log odds ratio models. You should specify either the LOGOR= or the TYPE= option, but not both.
Table 39.5 Log Odds Ratio Regression Structures Keyword
Log Odds Ratio Regression Structure
EXCH
Exchangeable
FULLCLUST
Fully parameterized clusters
LOGORVAR(variable)
Indicator variable for specifying block effects
NESTK
k-nested
NEST1
1-nested
ZFULL
Fully specified z matrix specified in ZDATA= data set
ZREP
Single cluster specification for replicated z matrix specified
in ZDATA= data set
ZREP(matrix)
Single cluster specification for replicated z matrix
- MAXITER=number
- MAXIT=number
specifies the maximum number of iterations allowed in the iterative GEE estimation process. The default number is 50.
- MCORRB
displays the estimated regression parameter model-based correlation matrix.
- MCOVB
displays the estimated regression parameter model-based covariance matrix.
- MODELSE
displays an analysis of parameter estimates table that uses model-based standard errors for inference. By default, an "Analysis of Parameter Estimates" table based on empirical standard errors is displayed.
- PRINTMLE
displays an analysis of maximum likelihood parameter estimates table. The maximum likelihood estimates are not displayed unless this option is specified.
- RUPDATE=number
specifies the number of iterations between updates of the working correlation matrix. For example, RUPDATE=5 specifies that the working correlation is updated once for every five regression parameter updates. The default value of number is 1; that is, the working correlation is updated every time the regression parameters are updated.
- SORTED
specifies that the input data are grouped by subject and sorted within subject. If this option is not specified, then the procedure internally sorts by subject-effect and within subject-effect, if a within subject-effect is specified.
- SUBCLUSTER=variable
- SUBCLUST=variable
specifies a variable defining subclusters for the 1-nested or k-nested log odds ratio association modeling structures. This variable must be listed in the CLASS statement.
- TYPE=correlation-structure keyword
- CORR=correlation-structure keyword
-
specifies the structure of the working correlation matrix used to model the correlation of the responses from subjects. Table 39.6 displays the correlation structure keywords and the corresponding correlation structures. The default working correlation type is the independent (CORR=IND). See the section Details: GENMOD Procedure for definitions of the correlation matrix types. You should specify LOGOR= or TYPE= but not both.
Table 39.6 Correlation Structure Types Keyword
Correlation Matrix Type
AR
AR(1)
Autoregressive(1)
EXCH
CS
Exchangeable
IND
Independent
MDEP(number)
 -dependent with
-dependent with  =number
=number UNSTR
UN
Unstructured
USER
FIXED (matrix)
Fixed, user-specified correlation matrix
For example, you can specify a fixed
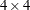 correlation matrix with the following option:
correlation matrix with the following option: TYPE=USER( 1.0 0.9 0.8 0.6 0.9 1.0 0.9 0.8 0.8 0.9 1.0 0.9 0.6 0.8 0.9 1.0 ) - V6CORR
specifies that the SAS ‘Version 6’ method of computing the normalized Pearson chi-square be used for working correlation estimation and for model-based covariance matrix scale factor.
- WITHINSUBJECT | WITHIN=within subject-effect
-
defines an effect specifying the order of measurements within subjects. Each distinct level of the within subject-effect defines a different response from the same subject. If the data are in proper order within each subject, you do not need to specify this option.
If some measurements do not appear in the data for some subjects, this option properly orders the existing measurements and treats the omitted measurements as missing values. If the WITHINSUBJECT= option is not used in this situation, measurements might be improperly ordered and missing values assumed for the last measurements in a cluster.
Variables used in defining the within subject-effect must be listed in the CLASS statement.
- YPAIR=variable-list
specifies the variables in the ZDATA= data set corresponding to pairs of responses for log odds ratio association modeling.
- ZDATA=SAS-data-set
specifies a SAS data set containing either the full z matrix for log odds ratio association modeling or the z matrix for a single complete cluster to be replicated for all clusters.
- ZROW=variable-list
specifies the variables in the ZDATA= data set corresponding to rows of the z matrix for log odds ratio association modeling.