The LIFEREG Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Missing Values Model Specification Computational Method Supported Distributions Predicted Values Confidence Intervals Fit Statistics Probability Plotting INEST= Data Set OUTEST= Data Set XDATA= Data Set Computational Resources Bayesian Analysis Displayed Output for Classical Analysis Displayed Output for Bayesian Analysis ODS Table Names ODS Graphics
Missing Values Model Specification Computational Method Supported Distributions Predicted Values Confidence Intervals Fit Statistics Probability Plotting INEST= Data Set OUTEST= Data Set XDATA= Data Set Computational Resources Bayesian Analysis Displayed Output for Classical Analysis Displayed Output for Bayesian Analysis ODS Table Names ODS Graphics -
Examples
 Motorette Failure Computing Predicted Values for a Tobit Model Overcoming Convergence Problems by Specifying Initial Values Analysis of Arbitrarily Censored Data with Interaction Effects Probability Plotting—Right Censoring Probability Plotting—Arbitrary Censoring Bayesian Analysis of Clinical Trial Data
Motorette Failure Computing Predicted Values for a Tobit Model Overcoming Convergence Problems by Specifying Initial Values Analysis of Arbitrarily Censored Data with Interaction Effects Probability Plotting—Right Censoring Probability Plotting—Arbitrary Censoring Bayesian Analysis of Clinical Trial Data - References
| Probability Plotting |
Probability plots are useful tools for the display and analysis of lifetime data. Probability plots use an inverse distribution scale so that a cumulative distribution function (CDF) plots as a straight line. A nonparametric estimate of the CDF of the lifetime data will plot approximately as a straight line, thus providing a visual assessment of goodness of fit.
You can use the PROBPLOT statement in PROC LIFEREG to create probability plots of data that are complete, right censored, interval censored, or a combination of censoring types (arbitrarily censored). A line representing the maximum likelihood fit from the MODEL statement and pointwise parametric confidence bands for the cumulative probabilities are also included in the plot.
A random variable 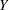 belongs to a location-scale family of distributions if its CDF
belongs to a location-scale family of distributions if its CDF 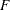 is of the form
is of the form
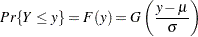 |
where 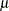 is the location parameter and
is the location parameter and  is the scale parameter. Here,
is the scale parameter. Here,  is a CDF that cannot depend on any unknown parameters, and
is a CDF that cannot depend on any unknown parameters, and  is the CDF of
is the CDF of 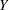 if
if 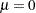 and
and  . For example, if
. For example, if 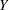 is a normal random variable with mean
is a normal random variable with mean 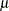 and standard deviation
and standard deviation  ,
,
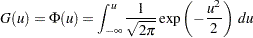 |
and
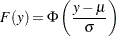 |
The normal, extreme-value, and logistic distributions are location-scale models. The three-parameter gamma distribution is a location-scale model if the shape parameter 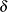 is fixed. If
is fixed. If 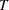 has a lognormal, Weibull, or log-logistic distribution, then
has a lognormal, Weibull, or log-logistic distribution, then 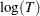 has a distribution that is a location-scale model. These distributions are said to be of type log-location-scale. Probability plots are constructed for lognormal, Weibull, and log-logistic distributions by using
has a distribution that is a location-scale model. These distributions are said to be of type log-location-scale. Probability plots are constructed for lognormal, Weibull, and log-logistic distributions by using 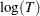 instead of
instead of 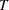 in the plots.
in the plots.
Let 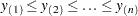 be ordered observations of a random sample with distribution function
be ordered observations of a random sample with distribution function 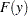 . A probability plot is a plot of the points
. A probability plot is a plot of the points 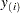 against
against 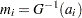 , where
, where 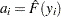 is an estimate of the CDF
is an estimate of the CDF 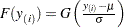 . The nonparametric CDF estimates
. The nonparametric CDF estimates 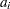 are sometimes called plotting positions. The axis on which the points
are sometimes called plotting positions. The axis on which the points 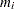 are plotted is usually labeled with a probability scale (the scale of
are plotted is usually labeled with a probability scale (the scale of 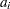 ).
).
If 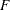 is one of the location-scale distributions, then
is one of the location-scale distributions, then 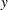 is the lifetime; otherwise, the log of the lifetime is used to transform the distribution to a location-scale model.
is the lifetime; otherwise, the log of the lifetime is used to transform the distribution to a location-scale model.
If the data actually have the stated distribution, then 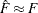 ,
,
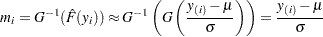 |
and points 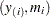 should fall approximately in a straight line.
should fall approximately in a straight line.
There are several ways to compute the nonparametric CDF estimates used in probability plots from lifetime data. These are discussed in the next two sections.
Complete and Right-Censored Data
The censoring times must be taken into account when you compute plotting positions for right-censored data. The modified Kaplan-Meier method described in the following section is the default method for computing nonparametric CDF estimates for display on probability plots. Refer to Abernethy (1996), Meeker and Escobar (1998), and Nelson (1982) for discussions of the methods described in the following sections.
Expected Ranks, Kaplan-Meier, and Modified Kaplan-Meier Methods
Let 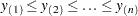 be ordered observations of a random sample including failure times and censor times. Order the data in increasing order. Label all the data with reverse ranks
be ordered observations of a random sample including failure times and censor times. Order the data in increasing order. Label all the data with reverse ranks  , with
, with 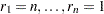 . For the lifetime (not censoring time) corresponding to reverse rank
. For the lifetime (not censoring time) corresponding to reverse rank  , compute the survival function estimate
, compute the survival function estimate
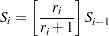 |
with 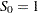 . The expected rank plotting position is computed as
. The expected rank plotting position is computed as 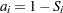 . The option PPOS=EXPRANK specifies the expected rank plotting position.
. The option PPOS=EXPRANK specifies the expected rank plotting position.
For the Kaplan-Meier method,
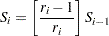 |
The Kaplan-Meier plotting position is then computed as 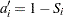 . The option PPOS=KM specifies the Kaplan-Meier plotting position.
. The option PPOS=KM specifies the Kaplan-Meier plotting position.
For the modified Kaplan-Meier method, use
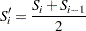 |
where 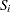 is computed from the Kaplan-Meier formula with
is computed from the Kaplan-Meier formula with 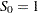 . The plotting position is then computed as
. The plotting position is then computed as 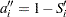 . The option PPOS=MKM specifies the modified Kaplan-Meier plotting position. If the PPOS option is not specified, the modified Kaplan-Meier plotting position is used as the default method.
. The option PPOS=MKM specifies the modified Kaplan-Meier plotting position. If the PPOS option is not specified, the modified Kaplan-Meier plotting position is used as the default method.
For complete samples, 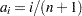 for the expected rank method,
for the expected rank method,  for the Kaplan-Meier method, and
for the Kaplan-Meier method, and 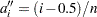 for the modified Kaplan-Meier method. If the largest observation is a failure for the Kaplan-Meier estimator, then
for the modified Kaplan-Meier method. If the largest observation is a failure for the Kaplan-Meier estimator, then  and the point is not plotted.
and the point is not plotted.
Median Ranks
Let 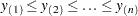 be ordered observations of a random sample including failure times and censor times. A failure order number
be ordered observations of a random sample including failure times and censor times. A failure order number 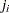 is assigned to the ith failure:
is assigned to the ith failure: 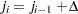 , where
, where 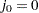 . The increment
. The increment 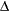 is initially 1 and is modified when a censoring time is encountered in the ordered sample. The new increment is computed as
is initially 1 and is modified when a censoring time is encountered in the ordered sample. The new increment is computed as
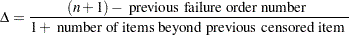 |
The plotting position is computed for the 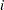 th failure time as
th failure time as
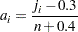 |
For complete samples, the failure order number 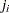 is equal to
is equal to 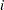 , the order of the failure in the sample. In this case, the preceding equation for
, the order of the failure in the sample. In this case, the preceding equation for 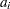 is an approximation of the median plotting position computed as the median of the ith-order statistic from the uniform distribution on (0, 1). In the censored case,
is an approximation of the median plotting position computed as the median of the ith-order statistic from the uniform distribution on (0, 1). In the censored case, 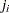 is not necessarily an integer, but the preceding equation still provides an approximation to the median plotting position. The PPOS=MEDRANK option specifies the median rank plotting position.
is not necessarily an integer, but the preceding equation still provides an approximation to the median plotting position. The PPOS=MEDRANK option specifies the median rank plotting position.
Arbitrarily Censored Data
The LIFEREG procedure can create probability plots for data that consist of combinations of exact, left-censored, right-censored, and interval-censored lifetimes—that is, arbitrarily censored data. The LIFEREG procedure uses an iterative algorithm developed by Turnbull (1976) to compute a nonparametric maximum likelihood estimate of the cumulative distribution function for the data. Since the technique is maximum likelihood, standard errors of the cumulative probability estimates are computed from the inverse of the associated Fisher information matrix. This algorithm is an example of the expectation-maximization (EM) algorithm. The default initial estimate assigns equal probabilities to each interval. You can specify different initial values with the PROBLIST= option. Convergence is determined if the change in the log likelihood between two successive iterations is less than delta, where the default value of delta is  . You can specify a different value for delta with the TOLLIKE= option. Iterations will be terminated if the algorithm does not converge after a fixed number of iterations. The default maximum number of iterations is 1000. Some data might require more iterations for convergence. You can specify the maximum allowed number of iterations with the MAXITEM= option in the PROBPLOT statement. The iteration history of the log likelihood is displayed if you specify the ITPRINTEM option. The iteration history of the estimated interval probabilities are also displayed if you specify both options ITPRINTEM and PRINTPROBS.
. You can specify a different value for delta with the TOLLIKE= option. Iterations will be terminated if the algorithm does not converge after a fixed number of iterations. The default maximum number of iterations is 1000. Some data might require more iterations for convergence. You can specify the maximum allowed number of iterations with the MAXITEM= option in the PROBPLOT statement. The iteration history of the log likelihood is displayed if you specify the ITPRINTEM option. The iteration history of the estimated interval probabilities are also displayed if you specify both options ITPRINTEM and PRINTPROBS.
If an interval probability is smaller than a tolerance ( by default) after convergence, the probability is set to zero, the interval probabilities are renormalized so that they add to one, and iterations are restarted. Usually the algorithm converges in just a few more iterations. You can change the default value of the tolerance with the TOLPROB= option. You can specify the NOPOLISH option to avoid setting small probabilities to zero and restarting the algorithm.
by default) after convergence, the probability is set to zero, the interval probabilities are renormalized so that they add to one, and iterations are restarted. Usually the algorithm converges in just a few more iterations. You can change the default value of the tolerance with the TOLPROB= option. You can specify the NOPOLISH option to avoid setting small probabilities to zero and restarting the algorithm.
If you specify the ITPRINTEM option, a table summarizing the Turnbull estimate of the interval probabilities is displayed. The columns labeled "Reduced Gradient" and "Lagrange Multiplier" are used in checking final convergence of the maximum likelihood estimate. The Lagrange multipliers must all be greater than or equal to zero, or the solution is not maximum likelihood. Refer to Gentleman and Geyer (1994) for more details of the convergence checking. Also refer to Meeker and Escobar (1998, Chapter 3) for more information.
See Example 50.6 for an illustration.
Nonparametric Confidence Intervals
You can use the PPOUT option in the PROBPLOT statement to create a table containing the nonparametric CDF estimates computed by the selected method, Kaplan-Meier CDF estimates, standard errors of the Kaplan-Meier estimator, and nonparametric confidence limits for the CDF. The confidence limits are either pointwise or simultaneous, depending on the value of the NPINTERVALS= option in the PROBPLOT statement. The method used in the LIFEREG procedure for computation of approximate pointwise and simultaneous confidence intervals for cumulative failure probabilities relies on the Kaplan-Meier estimator of the cumulative distribution function of failure time and approximate standard deviation of the Kaplan-Meier estimator. For the case of arbitrarily censored data, the Turnbull algorithm, discussed previously, provides an extension of the Kaplan-Meier estimator. Both the Kaplan-Meier and the Turnbull estimators provide an estimate of the standard error of the CDF estimator,  , that is used in computing confidence intervals.
, that is used in computing confidence intervals.
Pointwise Confidence Intervals
Approximate 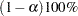 pointwise confidence intervals are computed as in Meeker and Escobar (1998, Section 3.6) as
pointwise confidence intervals are computed as in Meeker and Escobar (1998, Section 3.6) as
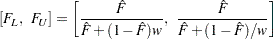 |
where
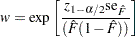 |
where 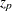 is the
is the 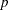 th quantile of the standard normal distribution.
th quantile of the standard normal distribution.
Simultaneous Confidence Intervals
Approximate 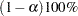 simultaneous confidence bands valid over the lifetime interval
simultaneous confidence bands valid over the lifetime interval 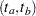 are computed as the "Equal Precision" case of Nair (1984) and Meeker and Escobar (1998, Section 3.8) as
are computed as the "Equal Precision" case of Nair (1984) and Meeker and Escobar (1998, Section 3.8) as
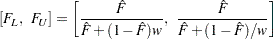 |
where
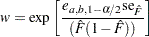 |
where the factor 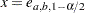 is the solution of
is the solution of
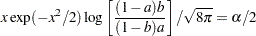 |
The time interval 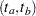 over which the bands are valid depends in a complicated way on the constants
over which the bands are valid depends in a complicated way on the constants  and
and 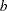 defined in Nair (1984),
defined in Nair (1984),  . The constants
. The constants  and
and 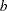 are chosen by default so that the confidence bands are valid between the lowest and highest times corresponding to failures in the case of multiply censored data, or to the lowest and highest intervals for which probabilities are computed for arbitrarily censored data. You can optionally specify
are chosen by default so that the confidence bands are valid between the lowest and highest times corresponding to failures in the case of multiply censored data, or to the lowest and highest intervals for which probabilities are computed for arbitrarily censored data. You can optionally specify  and
and 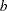 directly with the NPINTERVALS=SIMULTANEOUS(
directly with the NPINTERVALS=SIMULTANEOUS( ,
, 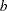 ) option in the PROBPLOT statement.
) option in the PROBPLOT statement.
Parametric Confidence Intervals
Pointwise parametric confidence bands are displayed in a probability plot, unless you specify the NOCONF option in the PROBPLOT statement. Two kinds of confidence intervals are available for display in a probability plot: confidence limits for the estimated cumulative distribution function (CDF) and confidence limits for estimated distribution percentiles.
Confidence Limits for the Estimated CDF
If the distribution is of type log-location-scale, let 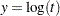 where
where  is the value of time at which the confidence limits are to be computed. If the distribution is of type location-scale, let
is the value of time at which the confidence limits are to be computed. If the distribution is of type location-scale, let 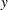 be the value at which you want to evaluate confidence limits for the estimated CDF
be the value at which you want to evaluate confidence limits for the estimated CDF 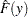 . Let
. Let
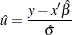 |
where the column vector  of covariate values is determined by the rules summarized in the section XDATA= Data Set. If an offset variable is specified, the mean of the offset variable values is included in
of covariate values is determined by the rules summarized in the section XDATA= Data Set. If an offset variable is specified, the mean of the offset variable values is included in 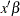 .
.
The CDF estimate is given by
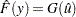 |
where  is the baseline distribution. The approximate standard error of
is the baseline distribution. The approximate standard error of 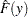 is computed as in Meeker and Escobar (1998, Section 8.4.3) as
is computed as in Meeker and Escobar (1998, Section 8.4.3) as
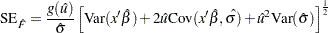 |
where 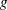 is the probability density function corresponding to
is the probability density function corresponding to  . Two-sided
. Two-sided 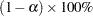 confidence limits are given by
confidence limits are given by
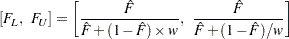 |
where
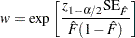 |
and 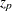 is the
is the 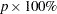 percentile of the standard normal distribution. The quantities
percentile of the standard normal distribution. The quantities 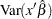 ,
, 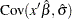 , and
, and 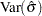 are computed based on the covariance matrix of the estimated parameter vector
are computed based on the covariance matrix of the estimated parameter vector 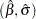 .
.
Confidence Limits for Percentiles
If the HCL option is specified in the PROBPLOT statement, confidence limits based on estimated distribution percentiles instead of the default CDF limits are displayed in the probability plot.
For location-scale distributions, the estimated 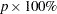 percentile of the distribution
percentile of the distribution 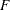 is given by
is given by
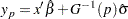 |
where  is the baseline distribution and the column vector
is the baseline distribution and the column vector  of covariate values is determined by the rules summarized in the section XDATA= Data Set. The standard error of
of covariate values is determined by the rules summarized in the section XDATA= Data Set. The standard error of 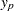 is estimated by
is estimated by 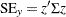 where
where 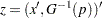 and
and 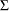 is the covariance matrix of the parameter estimates
is the covariance matrix of the parameter estimates  . Two-sided
. Two-sided 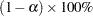 confidence limits for
confidence limits for 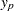 are given by
are given by
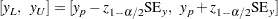 |
For distributions of type log-location-scale, the confidence limits are computed as
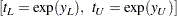 |
For example, if 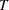 has the Weibull distribution,
has the Weibull distribution,  is the standardized extreme value distribution,
is the standardized extreme value distribution, 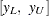 are confidence limits for the
are confidence limits for the 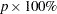 percentile of the extreme value distribution for
percentile of the extreme value distribution for 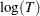 , and
, and 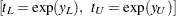 are confidence limits for the
are confidence limits for the 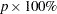 percentile of the Weibull distribution for
percentile of the Weibull distribution for 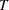 .
.