The HPSEVERITY Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Predefined DistributionsCensoring and TruncationParameter Estimation MethodDetails of Optimization AlgorithmsParameter InitializationEstimating Regression EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDistributed and Multithreaded ComputationDefining a Severity Distribution Model with the FCMP ProcedurePredefined Utility FunctionsCustom Objective FunctionsInput Data SetsOutput Data SetsDisplayed Output
Predefined DistributionsCensoring and TruncationParameter Estimation MethodDetails of Optimization AlgorithmsParameter InitializationEstimating Regression EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDistributed and Multithreaded ComputationDefining a Severity Distribution Model with the FCMP ProcedurePredefined Utility FunctionsCustom Objective FunctionsInput Data SetsOutput Data SetsDisplayed Output -
Examples
 Defining a Model for Gaussian DistributionDefining a Model for the Gaussian Distribution with a Scale ParameterDefining a Model for Mixed-Tail DistributionsFitting a Scaled Tweedie Model with RegressorsFitting Distributions to Interval-Censored DataBenefits of Distributed and Multithreaded ComputingEstimating Parameters Using Cramér-von Mises Estimator
Defining a Model for Gaussian DistributionDefining a Model for the Gaussian Distribution with a Scale ParameterDefining a Model for Mixed-Tail DistributionsFitting a Scaled Tweedie Model with RegressorsFitting Distributions to Interval-Censored DataBenefits of Distributed and Multithreaded ComputingEstimating Parameters Using Cramér-von Mises Estimator - References
A severity distribution model consists of a set of functions and subroutines that are defined using the FCMP procedure. The FCMP procedure is part of Base
SAS software. Each function or subroutine must be named as ![]() distribution-name
distribution-name![]() _
_![]() keyword
keyword![]() , where distribution-name is the identifying short name of the distribution and keyword identifies one of the functions or subroutines. The total length of the name should not exceed 32. Each function or subroutine
must have a specific signature, which consists of the number of arguments, sequence and types of arguments, and return value
type. The summary of all the recognized function and subroutine names and their expected behavior is given in Table 5.6.
, where distribution-name is the identifying short name of the distribution and keyword identifies one of the functions or subroutines. The total length of the name should not exceed 32. Each function or subroutine
must have a specific signature, which consists of the number of arguments, sequence and types of arguments, and return value
type. The summary of all the recognized function and subroutine names and their expected behavior is given in Table 5.6.
Consider the following points when you define a distribution model:
-
When you define a function or subroutine requiring parameter arguments, the names and order of those arguments must be the same. Arguments other than the parameter arguments can have any name, but they must satisfy the requirements on their type and order.
-
When the HPSEVERITY procedure invokes any function or subroutine, it provides the necessary input values according to the specified signature, and expects the function or subroutine to prepare the output and return it according to the specification of the return values in the signature.
-
You can use most of the SAS programming statements and SAS functions that you can use in a DATA step for defining the FCMP functions and subroutines. However, there are a few differences in the capabilities of the DATA step and the FCMP procedure. To learn more, see the documentation of the FCMP procedure in the Base SAS Procedures Guide.
-
You must specify either the PDF or the LOGPDF function. Similarly, you must specify either the CDF or the LOGCDF function. All other functions are optional, except when necessary for correct definition of the distribution. It is strongly recommended that you define the PARMINIT subroutine to provide a good set of initial values for the parameters. The information provided by PROC HPSEVERITY to the PARMINIT subroutine enables you to use popular initialization approaches based on the method of moments and the method of percentile matching, but you can implement any algorithm to initialize the parameters by using the values of the response variable and the estimate of its empirical distribution function.
-
The LOWERBOUNDS subroutines should be defined if the lower bound on at least one distribution parameter is different from the default lower bound of 0. If you define a LOWERBOUNDS subroutine but do not set a lower bound for some parameter inside the subroutine, then that parameter is assumed to have no lower bound (or a lower bound of
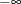 ). Hence, it is recommended that you explicitly return the lower bound for each parameter when you define the LOWERBOUNDS
subroutine.
). Hence, it is recommended that you explicitly return the lower bound for each parameter when you define the LOWERBOUNDS
subroutine.
-
The UPPERBOUNDS subroutines should be defined if the upper bound on at least one distribution parameter is different from the default upper bound of
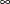 . If you define an UPPERBOUNDS subroutine but do not set an upper bound for some parameter inside the subroutine, then that
parameter is assumed to have no upper bound (or a upper bound of
. If you define an UPPERBOUNDS subroutine but do not set an upper bound for some parameter inside the subroutine, then that
parameter is assumed to have no upper bound (or a upper bound of 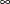 ). Hence, it is recommended that you explicitly return the upper bound for each parameter when you define the UPPERBOUNDS
subroutine.
). Hence, it is recommended that you explicitly return the upper bound for each parameter when you define the UPPERBOUNDS
subroutine.
-
If you want to use the distribution in a model with regression effects, then make sure that the first parameter of the distribution is the scale parameter itself or a log-transformed scale parameter. If the first parameter is a log-transformed scale parameter, then you must define the SCALETRANSFORM function.
-
In general, it is not necessary to define the gradient and Hessian functions, because the HPSEVERITY procedure uses an internal system to evaluate the required derivatives. The internal system typically computes the derivatives analytically. But it might not be able to do so if your function definitions use other functions that it cannot differentiate analytically. In such cases, derivatives are approximated using a finite difference method and a note is written to the SAS log to indicate the components that are differentiated using such approximations. PROC HPSEVERITY does reasonably well with these finite difference approximations. But, if you know of a way to compute the derivatives of such components analytically, then you should define the gradient and Hessian functions.
In order to use your distribution with PROC HPSEVERITY, you need to record the FCMP library that contains the functions and subroutines for your distribution and other FCMP libraries that contain FCMP functions or subroutines used within your distribution’s functions and subroutines. Specify all those libraries in the CMPLIB= system option by using the OPTIONS global statement. For more information about the OPTIONS statement, see SAS Statements: Reference. For more information about the CMPLIB= system option, see SAS System Options: Reference.
Each predefined distribution mentioned in the section Predefined Distributions has a distribution model associated with it. The functions and subroutines of all those models are available in the Sashelp.Svrtdist library. The order of the parameters in the signatures of the functions and subroutines is the same as listed in Table 5.2. You do not need to use the CMPLIB= option in order to use the predefined distributions with PROC HPSEVERITY. However, if
you need to use the functions or subroutines of the predefined distributions in SAS statements other than the PROC HPSEVERITY
step (such as in a DATA step), then specify the Sashelp.Svrtdist library in the CMPLIB= system option by using the OPTIONS global statement prior to using them.
Table 5.6 shows functions and subroutines that define a distribution model, and subsections after the table provide more detail. The functions are listed in alphabetical order of the keyword suffix.
Table 5.6: List of Functions and Subroutines That Define a Distribution Model
|
Name |
Type |
Required |
Expected to Return |
|---|---|---|---|
|
Function |
YES |
Cumulative distribution |
|
|
function value |
|||
|
Subroutine |
NO |
Gradient of the CDF |
|
|
Subroutine |
NO |
Hessian of the CDF |
|
|
Subroutine |
NO |
Constant parameters |
|
|
Function |
NO |
Description of the distribution |
|
|
Function |
YES |
Log of cumulative distribution |
|
|
function value |
|||
|
Subroutine |
NO |
Gradient of the LOGCDF |
|
|
Subroutine |
NO |
Hessian of the LOGCDF |
|
|
Function |
YES |
Log of probability density |
|
|
function value |
|||
|
Subroutine |
NO |
Gradient of the LOGPDF |
|
|
Subroutine |
NO |
Hessian of the LOGPDF |
|
|
Function |
NO |
Log of survival |
|
|
function value |
|||
|
Subroutine |
NO |
Gradient of the LOGSDF |
|
|
Subroutine |
NO |
Hessian of the LOGSDF |
|
|
Subroutine |
NO |
Lower bounds on parameters |
|
|
Subroutine |
NO |
Initial values |
|
|
for parameters |
|||
|
Function |
YES |
Probability density |
|
|
function value |
|||
|
Subroutine |
NO |
Gradient of the PDF |
|
|
Subroutine |
NO |
Hessian of the PDF |
|
|
Function |
NO |
Type of relationship between |
|
|
the first distribution parameter |
|||
|
and the scale parameter |
|||
|
Function |
NO |
Survival function value |
|
|
Subroutine |
NO |
Gradient of the SDF |
|
|
Subroutine |
NO |
Hessian of the SDF |
|
|
Subroutine |
NO |
Upper bounds on parameters |
|
|
Notes: |
|||
|
1. Either the dist_CDF or the dist_LOGCDF function must be defined. |
|||
|
2. Either the dist_PDF or the dist_LOGPDF function must be defined. |
|||
The signature syntax and semantics of each function or subroutine are as follows: