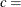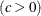The UNIVARIATE Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Missing ValuesRoundingDescriptive StatisticsCalculating the ModeCalculating PercentilesTests for LocationConfidence Limits for Parameters of the Normal DistributionRobust EstimatorsCreating Line Printer PlotsCreating High-Resolution GraphicsUsing the CLASS Statement to Create Comparative PlotsPositioning InsetsFormulas for Fitted Continuous DistributionsGoodness-of-Fit TestsKernel Density EstimatesConstruction of Quantile-Quantile and Probability PlotsInterpretation of Quantile-Quantile and Probability PlotsDistributions for Probability and Q-Q PlotsEstimating Shape Parameters Using Q-Q PlotsEstimating Location and Scale Parameters Using Q-Q PlotsEstimating Percentiles Using Q-Q PlotsInput Data SetsOUT= Output Data Set in the OUTPUT StatementOUTHISTOGRAM= Output Data SetOUTKERNEL= Output Data SetOUTTABLE= Output Data SetTables for Summary StatisticsODS Table NamesODS Tables for Fitted DistributionsODS GraphicsComputational Resources
Missing ValuesRoundingDescriptive StatisticsCalculating the ModeCalculating PercentilesTests for LocationConfidence Limits for Parameters of the Normal DistributionRobust EstimatorsCreating Line Printer PlotsCreating High-Resolution GraphicsUsing the CLASS Statement to Create Comparative PlotsPositioning InsetsFormulas for Fitted Continuous DistributionsGoodness-of-Fit TestsKernel Density EstimatesConstruction of Quantile-Quantile and Probability PlotsInterpretation of Quantile-Quantile and Probability PlotsDistributions for Probability and Q-Q PlotsEstimating Shape Parameters Using Q-Q PlotsEstimating Location and Scale Parameters Using Q-Q PlotsEstimating Percentiles Using Q-Q PlotsInput Data SetsOUT= Output Data Set in the OUTPUT StatementOUTHISTOGRAM= Output Data SetOUTKERNEL= Output Data SetOUTTABLE= Output Data SetTables for Summary StatisticsODS Table NamesODS Tables for Fitted DistributionsODS GraphicsComputational Resources -
Examples
 Computing Descriptive Statistics for Multiple VariablesCalculating ModesIdentifying Extreme Observations and Extreme ValuesCreating a Frequency TableCreating Basic Summary PlotsAnalyzing a Data Set With a FREQ VariableSaving Summary Statistics in an OUT= Output Data SetSaving Percentiles in an Output Data SetComputing Confidence Limits for the Mean, Standard Deviation, and VarianceComputing Confidence Limits for Quantiles and PercentilesComputing Robust EstimatesTesting for LocationPerforming a Sign Test Using Paired DataCreating a HistogramCreating a One-Way Comparative HistogramCreating a Two-Way Comparative HistogramAdding Insets with Descriptive StatisticsBinning a HistogramAdding a Normal Curve to a HistogramAdding Fitted Normal Curves to a Comparative HistogramFitting a Beta CurveFitting Lognormal, Weibull, and Gamma CurvesComputing Kernel Density EstimatesFitting a Three-Parameter Lognormal CurveAnnotating a Folded Normal CurveCreating Lognormal Probability PlotsCreating a Histogram to Display Lognormal FitCreating a Normal Quantile PlotAdding a Distribution Reference LineInterpreting a Normal Quantile PlotEstimating Three Parameters from Lognormal Quantile PlotsEstimating Percentiles from Lognormal Quantile PlotsEstimating Parameters from Lognormal Quantile PlotsComparing Weibull Quantile PlotsCreating a Cumulative Distribution PlotCreating a P-P Plot
Computing Descriptive Statistics for Multiple VariablesCalculating ModesIdentifying Extreme Observations and Extreme ValuesCreating a Frequency TableCreating Basic Summary PlotsAnalyzing a Data Set With a FREQ VariableSaving Summary Statistics in an OUT= Output Data SetSaving Percentiles in an Output Data SetComputing Confidence Limits for the Mean, Standard Deviation, and VarianceComputing Confidence Limits for Quantiles and PercentilesComputing Robust EstimatesTesting for LocationPerforming a Sign Test Using Paired DataCreating a HistogramCreating a One-Way Comparative HistogramCreating a Two-Way Comparative HistogramAdding Insets with Descriptive StatisticsBinning a HistogramAdding a Normal Curve to a HistogramAdding Fitted Normal Curves to a Comparative HistogramFitting a Beta CurveFitting Lognormal, Weibull, and Gamma CurvesComputing Kernel Density EstimatesFitting a Three-Parameter Lognormal CurveAnnotating a Folded Normal CurveCreating Lognormal Probability PlotsCreating a Histogram to Display Lognormal FitCreating a Normal Quantile PlotAdding a Distribution Reference LineInterpreting a Normal Quantile PlotEstimating Three Parameters from Lognormal Quantile PlotsEstimating Percentiles from Lognormal Quantile PlotsEstimating Parameters from Lognormal Quantile PlotsComparing Weibull Quantile PlotsCreating a Cumulative Distribution PlotCreating a P-P Plot - References
PPPLOT Statement
-
PPPLOT <variables> < / options>;
The PPPLOT statement creates a probability-probability plot (also referred to as a P-P plot or percent plot), which compares the empirical cumulative distribution function (ECDF) of a variable with a specified theoretical cumulative distribution function such as the normal. If the two distributions match, the points on the plot form a linear pattern that passes through the origin and has unit slope. Thus, you can use a P-P plot to determine how well a theoretical distribution models a set of measurements.
You can specify one of the following theoretical distributions with the PPPLOT statement:
-
beta
-
exponential
-
gamma
-
Gumbel
-
generalized Pareto
-
inverse Gaussian
-
lognormal
-
normal
-
power function
-
Rayleigh
-
Weibull
Note: Probability-probability plots should not be confused with probability plots, which compare a set of ordered measurements with percentiles from a specified distribution. You can create probability plots with the PROBPLOT statement.
You can use any number of PPPLOT statements in the UNIVARIATE procedure. The components of the PPPLOT statement are as follows.
Distribution Options
Table 4.15 summarizes the options for requesting a specific theoretical distribution.
Table 4.15: Options for Specifying the Theoretical Distribution
|
Option |
Description |
|---|---|
|
specifies beta P-P plot |
|
|
specifies exponential P-P plot |
|
|
specifies gamma P-P plot |
|
|
specifies Gumbel P-P plot |
|
|
specifies generalized Pareto P-P plot |
|
|
specifies inverse Gaussian P-P plot |
|
|
specifies lognormal P-P plot |
|
|
specifies normal P-P plot |
|
|
specifies power function P-P plot |
|
|
specifies Rayleigh P-P plot |
|
|
specifies Weibull P-P plot |
Table 4.16 summarizes options that specify distribution parameters and control the display of the diagonal distribution reference line. Specify these options in parentheses after the distribution option. For example, the following statements use the NORMAL option to request a normal P-P plot:
proc univariate data=measures; ppplot length / normal(mu=10 sigma=0.3 color=red); run;
The MU=
and SIGMA=
normal-options specify 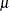 and
and  for the normal distribution, and the COLOR=
normal-option specifies the color for the line.
for the normal distribution, and the COLOR=
normal-option specifies the color for the line.
Table 4.16: Secondary Distribution Reference Line Options
|
Option |
Description |
|---|---|
|
Options Used with All Distributions |
|
|
specifies color of distribution reference line |
|
|
specifies line type of distribution reference line |
|
|
suppresses the distribution reference line |
|
|
specifies width of distribution reference line |
|
|
Beta-Options |
|
|
specifies shape parameter |
|
|
specifies shape parameter |
|
|
specifies scale parameter |
|
|
specifies lower threshold parameter |
|
|
Exponential-Options |
|
|
specifies scale parameter |
|
|
specifies threshold parameter |
|
|
Gamma-Options |
|
|
specifies shape parameter |
|
|
specifies change in successive estimates of |
|
|
specifies initial value for |
|
|
specifies maximum number of iterations in the Newton-Raphson approximation of |
|
|
specifies scale parameter |
|
|
specifies threshold parameter |
|
|
Gumbel-Options |
|
|
specifies location parameter |
|
|
specifies scale parameter |
|
|
IGauss-Options |
|
|
specifies shape parameter |
|
|
specifies mean |
|
|
Lognormal-Options |
|
|
specifies shape parameter |
|
|
specifies threshold parameter |
|
|
specifies scale parameter |
|
|
Normal-Options |
|
|
specifies mean |
|
|
specifies standard deviation |
|
|
Pareto-Options |
|
|
specifies shape parameter |
|
|
specifies scale parameter |
|
|
specifies threshold parameter |
|
|
Power-Options |
|
|
specifies shape parameter |
|
|
specifies scale parameter |
|
|
specifies threshold parameter |
|
|
Rayleigh-Options |
|
|
specifies scale parameter |
|
|
specifies threshold parameter |
|
|
Weibull-Options |
|
|
specifies shape parameter c |
|
|
requests table of iteration history and optimizer details |
|
|
specifies maximum number of iterations in the Newton-Raphson approximation of |
|
|
specifies scale parameter |
|
|
specifies threshold parameter |
|
General Options
Table 4.17 lists options that control the appearance of the plots. For complete descriptions, see the sections Dictionary of Options and Dictionary of Common Options.
Table 4.17: General Graphics Options
|
Option |
Description |
|---|---|
|
General Graphics Options |
|
|
specifies reference lines perpendicular to the horizontal axis |
|
|
specifies line labels for HREF= lines |
|
|
specifies position for HREF= line labels |
|
|
suppresses label for horizontal axis |
|
|
suppresses label for vertical axis |
|
|
suppresses tick marks and tick mark labels for vertical axis |
|
|
displays P-P plot in square format |
|
|
specifies label for vertical axis |
|
|
specifies reference lines perpendicular to the vertical axis |
|
|
specifies line labels for VREF= lines |
|
|
specifies position for VREF= line labels |
|
|
Options for Traditional Graphics Output |
|
|
provides an annotate data set |
|
|
specifies color for axis |
|
|
specifies color for frame |
|
|
specifies colors for HREF= lines |
|
|
specifies color for text |
|
|
specifies colors for VREF= lines |
|
|
specifies description for plot in graphics catalog |
|
|
specifies software font for text |
|
|
specifies AXIS statement for horizontal axis |
|
|
specifies height of text used outside framed areas |
|
|
specifies number of minor tick marks on horizontal axis |
|
|
specifies software font for text inside framed areas |
|
|
specifies height of text inside framed areas |
|
|
specifies line types for HREF= lines |
|
|
specifies line types for VREF= lines |
|
|
specifies name for plot in graphics catalog |
|
|
suppresses frame around plotting area |
|
|
turns and vertically strings out characters in labels for vertical axis |
|
|
specifies AXIS statement for vertical axis |
|
|
specifies number of minor tick marks on vertical axis |
|
|
specifies line thickness for axes and frame |
|
|
Options for ODS Graphics Output |
|
|
specifies footnote displayed on plot |
|
|
specifies secondary footnote displayed on plot |
|
|
specifies title displayed on plot |
|
|
specifies secondary title displayed on plot |
|
|
overlays plots for different class levels (ODS Graphics only) |
|
|
Options for Comparative Plots |
|
|
applies annotation requested in ANNOTATE= data set to key cell only |
|
|
specifies color for filling row label frames |
|
|
specifies color for filling column label frames |
|
|
specifies color for proportion of frequency bar |
|
|
specifies color for row labels |
|
|
specifies color for column labels |
|
|
specifies distance between tiles in comparative plot |
|
|
specifies number of columns in comparative plot |
|
|
specifies number of rows in comparative plot |
|
|
Miscellaneous Options |
|
|
specifies table of contents entry for P-P plot grouping |
|
Dictionary of Options
The following entries provide detailed descriptions of options for the PPPLOT statement. See the section Dictionary of Common Options for detailed descriptions of options common to all plot statements.

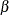
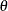
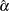
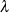
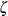
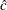
![\[ x_{(1)} \leq x_{(2)} \leq \cdots \leq x_{(n)} \]](images/procstat_univariate0081.png)
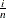
![\[ B_{\alpha \beta }\left(\frac{x_{(i)}-\theta }{\sigma }\right) = \int _{\theta }^{x_{(i)}} \frac{(t-\theta )^{\alpha -1}(\theta +\sigma -t)^{\beta -1} }{B(\alpha ,\beta ) \sigma ^{(\alpha +\beta -1)} } dt \]](images/procstat_univariate0083.png)
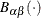
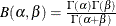
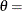
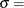
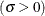
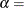
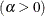
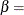
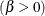
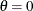
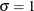
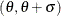
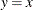
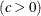
![\[ F(x_{(i)}) = 1-\exp \left(-\frac{x_{(i)}-\theta }{\sigma }\right) \]](images/procstat_univariate0095.png)
![\[ G_{\alpha }\left(\frac{x_{(i)}-\theta }{\sigma }\right) = \int _{\theta }^{x_{(i)}} \frac{1}{\sigma \Gamma (\alpha )} \left(\frac{t-\theta }{\sigma }\right)^{\alpha -1} \exp \left(-\frac{t-\theta }{\sigma }\right) dt \]](images/procstat_univariate0096.png)
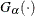
![\[ F\left(x_{(i)}\right) = \exp (-e^{-(x_{(i)}-\mu )/\sigma }) \]](images/procstat_univariate0098.png)
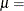
![\[ F(x_{(i)}) = \Phi \left\{ \sqrt {\frac{\lambda }{x_{(i)}}} \left( \frac{x_{(i)}}{\mu }-1\right) \right\} + e^{2\lambda /\mu } \Phi \left\{ -\sqrt {\frac{\lambda }{x_{(i)}}} \left( \frac{x_{(i)}}{\mu }+1\right) \right\} \]](images/procstat_univariate0100.png)
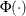
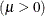
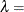
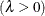
![\[ \Phi \left(\frac{\log (x_{(i)}-\theta )-\zeta }{\sigma }\right) \]](images/procstat_univariate0103.png)
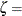
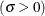
![\[ \Phi \left(\frac{x_{(i)}-\mu }{\sigma }\right) = \int _{-\infty }^{x_{(i)}} \frac{1}{\sigma \sqrt {2 \pi } } \exp \left( -\frac{(t - \mu )^2}{2 \sigma ^{2}} \right) dt \]](images/procstat_univariate0105.png)
![\[ F(x_{(i)}) = 1 - { \left( 1 - \frac{\alpha (x_{(i)} - \theta )}{\sigma } \right) }^\frac {1}{\alpha } \]](images/procstat_univariate0106.png)
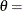
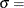
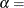
![\[ F(x_{(i)}) = {\left( \frac{x_{(i)} - \theta }{\sigma } \right)}^{\alpha } \]](images/procstat_univariate0107.png)
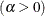
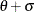
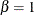
![\[ F(x_{(i)}) = 1 - e^{-(x_{(i)} - \theta )^2/(2\sigma ^2)} \]](images/procstat_univariate0108.png)
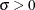
![\[ F(x_{(i)}) = 1-\exp \left( -\left( \frac{x_{(i)}-\theta }{\sigma } \right)^{c} \right) \]](images/procstat_univariate0110.png)