The NLMIXED Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 Modeling Assumptions and Notation Integral Approximations Built-in Log-Likelihood Functions Optimization Algorithms Finite-Difference Approximations of Derivatives Hessian Scaling Active Set Methods Line-Search Methods Restricting the Step Length Computational Problems Covariance Matrix Prediction Computational Resources Displayed Output ODS Table Names
Modeling Assumptions and Notation Integral Approximations Built-in Log-Likelihood Functions Optimization Algorithms Finite-Difference Approximations of Derivatives Hessian Scaling Active Set Methods Line-Search Methods Restricting the Step Length Computational Problems Covariance Matrix Prediction Computational Resources Displayed Output ODS Table Names -
Examples

- References
Example 63.2 Probit-Normal Model with Binomial Data
For this example, consider the data from Weil (1970), also studied by Williams (1975), Ochi and Prentice (1984), and McCulloch (1994). In this experiment 16 pregnant rats receive a control diet and 16 receive a chemically treated diet, and the litter size for each rat is recorded after 4 and 21 days. The SAS data set follows:
data rats;
input trt $ m x @@;
if (trt='c') then do;
x1 = 1;
x2 = 0;
end;
else do;
x1 = 0;
x2 = 1;
end;
litter = _n_;
datalines;
c 13 13 c 12 12 c 9 9 c 9 9 c 8 8 c 8 8 c 13 12 c 12 11
c 10 9 c 10 9 c 9 8 c 13 11 c 5 4 c 7 5 c 10 7 c 10 7
t 12 12 t 11 11 t 10 10 t 9 9 t 11 10 t 10 9 t 10 9 t 9 8
t 9 8 t 5 4 t 9 7 t 7 4 t 10 5 t 6 3 t 10 3 t 7 0
;
Here, m represents the size of the litter after 4 days, and x represents the size of the litter after 21 days. Also, indicator variables x1 and x2 are constructed for the two treatment levels.
Following McCulloch (1994), assume a latent survival model of the form
 |
where 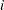 indexes treatment,
indexes treatment,  indexes litter, and
indexes litter, and 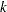 indexes newborn rats within a litter. The
indexes newborn rats within a litter. The 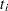 represent treatment means, the
represent treatment means, the 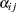 represent random litter effects assumed to be iid
represent random litter effects assumed to be iid 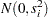 , and the
, and the 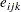 represent iid residual errors, all on the latent scale.
represent iid residual errors, all on the latent scale.
Instead of observing the survival times 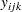 , assume that only the binary variable indicating whether
, assume that only the binary variable indicating whether 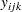 exceeds 0 is observed. If
exceeds 0 is observed. If  denotes the sum of these binary variables for the
denotes the sum of these binary variables for the 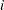 th treatment and the
th treatment and the  th litter, then the preceding assumptions lead to the following generalized linear mixed model:
th litter, then the preceding assumptions lead to the following generalized linear mixed model:
 |
where 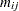 is the size of each litter after 4 days and
is the size of each litter after 4 days and
 |
The PROC NLMIXED statements to fit this model are as follows:
proc nlmixed data=rats; parms t1=1 t2=1 s1=.05 s2=1; eta = x1*t1 + x2*t2 + alpha; p = probnorm(eta); model x ~ binomial(m,p); random alpha ~ normal(0,x1*s1*s1+x2*s2*s2) subject=litter; estimate 'gamma2' t2/sqrt(1+s2*s2); predict p out=p; run;
As in Example 63.1, the PROC NLMIXED statement invokes the procedure and the PARMS statement defines the parameters. The parameters for this example are the two treatment means, t1 and t2, and the two random-effect standard deviations, s1 and s2.
The indicator variables x1 and x2 are used in the program to assign the proper mean to each observation in the input data set as well as the proper variance to the random effects. Note that programming expressions are permitted inside the distributional specifications, as illustrated by the random-effects variance specified here.
The ESTIMATE statement requests an estimate of 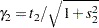 , which is a location-scale parameter from Ochi and Prentice (1984).
, which is a location-scale parameter from Ochi and Prentice (1984).
The PREDICT statement constructs predictions for each observation in the input data set. For this example, predictions of p and approximate standard errors of prediction are output to a SAS data set named P. These predictions are functions of the parameter estimates and the empirical Bayes estimates of the random effects 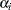 .
.
The output for this model is as follows.
| Specifications | |
|---|---|
| Data Set | WORK.RATS |
| Dependent Variable | x |
| Distribution for Dependent Variable | Binomial |
| Random Effects | alpha |
| Distribution for Random Effects | Normal |
| Subject Variable | litter |
| Optimization Technique | Dual Quasi-Newton |
| Integration Method | Adaptive Gaussian Quadrature |
| Dimensions | |
|---|---|
| Observations Used | 32 |
| Observations Not Used | 0 |
| Total Observations | 32 |
| Subjects | 32 |
| Max Obs Per Subject | 1 |
| Parameters | 4 |
| Quadrature Points | 7 |
| Parameters | ||||
|---|---|---|---|---|
| t1 | t2 | s1 | s2 | NegLogLike |
| 1 | 1 | 0.05 | 1 | 54.9362323 |
The "Specifications" table provides basic information about this nonlinear mixed model (Output 63.2.1). The "Dimensions" table provides counts of various variables. Note that each observation in the data comprises a separate subject. Using the starting values in the "Parameters" table, PROC NLMIXED determines that the log-likelihood function can be approximated with sufficient accuracy with a seven-point quadrature rule.
| Iteration History | ||||||
|---|---|---|---|---|---|---|
| Iter | Calls | NegLogLike | Diff | MaxGrad | Slope | |
| 1 | 2 | 53.9933934 | 0.942839 | 11.03261 | -81.9428 | |
| 2 | 3 | 52.875353 | 1.11804 | 2.148952 | -2.86277 | |
| 3 | 5 | 52.6350386 | 0.240314 | 0.329957 | -1.05049 | |
| 4 | 6 | 52.6319939 | 0.003045 | 0.122926 | -0.00672 | |
| 5 | 8 | 52.6313583 | 0.000636 | 0.028246 | -0.00352 | |
| 6 | 11 | 52.6313174 | 0.000041 | 0.013551 | -0.00023 | |
| 7 | 13 | 52.6313115 | 5.839E-6 | 0.000603 | -0.00001 | |
| 8 | 15 | 52.6313115 | 9.45E-9 | 0.000022 | -1.68E-8 | |
| NOTE: GCONV convergence criterion satisfied. |
The "Iteration History" table indicates successful convergence in 8 iterations (Output 63.2.2).
| Fit Statistics | |
|---|---|
| -2 Log Likelihood | 105.3 |
| AIC (smaller is better) | 113.3 |
| AICC (smaller is better) | 114.7 |
| BIC (smaller is better) | 119.1 |
The "Fit Statistics" table lists useful statistics based on the maximized value of the log likelihood (Output 63.2.3).
| Parameter Estimates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Estimate | Standard Error | DF | t Value | Pr > |t| | Alpha | Lower | Upper | Gradient |
| t1 | 1.3063 | 0.1685 | 31 | 7.75 | <.0001 | 0.05 | 0.9626 | 1.6499 | -0.00002 |
| t2 | 0.9475 | 0.3055 | 31 | 3.10 | 0.0041 | 0.05 | 0.3244 | 1.5705 | 9.283E-6 |
| s1 | 0.2403 | 0.3015 | 31 | 0.80 | 0.4315 | 0.05 | -0.3746 | 0.8552 | 0.000014 |
| s2 | 1.0292 | 0.2988 | 31 | 3.44 | 0.0017 | 0.05 | 0.4198 | 1.6385 | -3.16E-6 |
The "Parameter Estimates" table indicates significance of all the parameters except S1 (Output 63.2.4).
| Additional Estimates | ||||||||
|---|---|---|---|---|---|---|---|---|
| Label | Estimate | Standard Error | DF | t Value | Pr > |t| | Alpha | Lower | Upper |
| gamma2 | 0.6603 | 0.2165 | 31 | 3.05 | 0.0047 | 0.05 | 0.2186 | 1.1019 |
The "Additional Estimates" table displays results from the ESTIMATE statement (Output 63.2.5). The estimate of  equals
equals 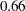 , agreeing with that obtained by McCulloch (1994). The standard error
, agreeing with that obtained by McCulloch (1994). The standard error 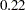 is computed using the delta method (Billingsley 1986; Cox, 1998).
is computed using the delta method (Billingsley 1986; Cox, 1998).
Not shown is the P data set, which contains the original 32 observations and predictions of the  .
.