The NLMIXED Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 Modeling Assumptions and Notation Integral Approximations Built-in Log-Likelihood Functions Optimization Algorithms Finite-Difference Approximations of Derivatives Hessian Scaling Active Set Methods Line-Search Methods Restricting the Step Length Computational Problems Covariance Matrix Prediction Computational Resources Displayed Output ODS Table Names
Modeling Assumptions and Notation Integral Approximations Built-in Log-Likelihood Functions Optimization Algorithms Finite-Difference Approximations of Derivatives Hessian Scaling Active Set Methods Line-Search Methods Restricting the Step Length Computational Problems Covariance Matrix Prediction Computational Resources Displayed Output ODS Table Names -
Examples

- References
Example 63.1 One-Compartment Model with Pharmacokinetic Data
A popular application of nonlinear mixed models is in the field of pharmacokinetics, which studies how a drug disperses through a living individual. This example considers the theophylline data from Pinheiro and Bates (1995). Serum concentrations of the drug theophylline are measured in 12 subjects over a 25-hour period after oral administration. The data are as follows.
data theoph; input subject time conc dose wt; datalines; 1 0.00 0.74 4.02 79.6 1 0.25 2.84 4.02 79.6 1 0.57 6.57 4.02 79.6 1 1.12 10.50 4.02 79.6 1 2.02 9.66 4.02 79.6 1 3.82 8.58 4.02 79.6 1 5.10 8.36 4.02 79.6 ... more lines ... 12 24.15 1.17 5.30 60.5 ;
Pinheiro and Bates (1995) consider the following first-order compartment model for these data:
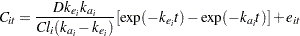 |
where 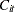 is the observed concentration of the
is the observed concentration of the 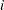 th subject at time
th subject at time  ,
, 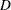 is the dose of theophylline,
is the dose of theophylline, 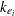 is the elimination rate constant for subject
is the elimination rate constant for subject 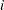 ,
, 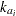 is the absorption rate constant for subject
is the absorption rate constant for subject 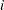 ,
, 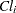 is the clearance for subject
is the clearance for subject 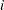 , and
, and 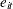 are normal errors. To allow for random variability between subjects, they assume
are normal errors. To allow for random variability between subjects, they assume
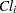 |
 |
|||
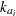 |
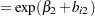 |
|||
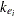 |
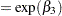 |
where the 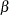 s denote fixed-effects parameters and the
s denote fixed-effects parameters and the 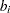 s denote random-effects parameters with an unknown covariance matrix.
s denote random-effects parameters with an unknown covariance matrix.
The PROC NLMIXED statements to fit this model are as follows:
proc nlmixed data=theoph;
parms beta1=-3.22 beta2=0.47 beta3=-2.45
s2b1 =0.03 cb12 =0 s2b2 =0.4 s2=0.5;
cl = exp(beta1 + b1);
ka = exp(beta2 + b2);
ke = exp(beta3);
pred = dose*ke*ka*(exp(-ke*time)-exp(-ka*time))/cl/(ka-ke);
model conc ~ normal(pred,s2);
random b1 b2 ~ normal([0,0],[s2b1,cb12,s2b2]) subject=subject;
run;
The PARMS statement specifies starting values for the three 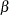 s and four variance-covariance parameters. The clearance and rate constants are defined using SAS programming statements, and the conditional model for the data is defined to be normal with mean pred and variance s2. The two random effects are b1 and b2, and their joint distribution is defined in the RANDOM statement. Brackets are used in defining their mean vector (two zeros) and the lower triangle of their variance-covariance matrix (a general
s and four variance-covariance parameters. The clearance and rate constants are defined using SAS programming statements, and the conditional model for the data is defined to be normal with mean pred and variance s2. The two random effects are b1 and b2, and their joint distribution is defined in the RANDOM statement. Brackets are used in defining their mean vector (two zeros) and the lower triangle of their variance-covariance matrix (a general 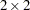 matrix). The SUBJECT= variable is subject.
matrix). The SUBJECT= variable is subject.
The results from this analysis are as follows.
| Specifications | |
|---|---|
| Data Set | WORK.THEOPH |
| Dependent Variable | conc |
| Distribution for Dependent Variable | Normal |
| Random Effects | b1 b2 |
| Distribution for Random Effects | Normal |
| Subject Variable | subject |
| Optimization Technique | Dual Quasi-Newton |
| Integration Method | Adaptive Gaussian Quadrature |
The "Specifications" table lists the setup of the model (Output 63.1.1). The "Dimensions" table indicates that there are 132 observations, 12 subjects, and 7 parameters. PROC NLMIXED selects 5 quadrature points for each random effect, producing a total grid of 25 points over which quadrature is performed (Output 63.1.2).
| Dimensions | |
|---|---|
| Observations Used | 132 |
| Observations Not Used | 0 |
| Total Observations | 132 |
| Subjects | 12 |
| Max Obs Per Subject | 11 |
| Parameters | 7 |
| Quadrature Points | 5 |
The "Parameters" table lists the 7 parameters, their starting values, and the initial evaluation of the negative log likelihood using adaptive Gaussian quadrature (Output 63.1.3). The "Iteration History" table indicates that 10 steps are required for the dual quasi-Newton algorithm to achieve convergence.
| Parameters | |||||||
|---|---|---|---|---|---|---|---|
| beta1 | beta2 | beta3 | s2b1 | cb12 | s2b2 | s2 | NegLogLike |
| -3.22 | 0.47 | -2.45 | 0.03 | 0 | 0.4 | 0.5 | 177.789945 |
| Iteration History | ||||||
|---|---|---|---|---|---|---|
| Iter | Calls | NegLogLike | Diff | MaxGrad | Slope | |
| 1 | 5 | 177.776248 | 0.013697 | 2.873367 | -63.0744 | |
| 2 | 8 | 177.7643 | 0.011948 | 1.698144 | -4.75239 | |
| 3 | 10 | 177.757264 | 0.007036 | 1.297439 | -1.97311 | |
| 4 | 12 | 177.755688 | 0.001576 | 1.441408 | -0.49772 | |
| 5 | 14 | 177.7467 | 0.008988 | 1.132279 | -0.8223 | |
| 6 | 17 | 177.746401 | 0.000299 | 0.831293 | -0.00244 | |
| 7 | 19 | 177.746318 | 0.000083 | 0.724198 | -0.00789 | |
| 8 | 21 | 177.74574 | 0.000578 | 0.180018 | -0.00583 | |
| 9 | 23 | 177.745736 | 3.88E-6 | 0.017958 | -8.25E-6 | |
| 10 | 25 | 177.745736 | 3.222E-8 | 0.000143 | -6.51E-8 | |
| NOTE: GCONV convergence criterion satisfied. |
| Fit Statistics | |
|---|---|
| -2 Log Likelihood | 355.5 |
| AIC (smaller is better) | 369.5 |
| AICC (smaller is better) | 370.4 |
| BIC (smaller is better) | 372.9 |
The "Fit Statistics" table lists the final optimized values of the log-likelihood function and information criteria in the "smaller is better" form (Output 63.1.4).
| Parameter Estimates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Estimate | Standard Error | DF | t Value | Pr > |t| | Alpha | Lower | Upper | Gradient |
| beta1 | -3.2268 | 0.05950 | 10 | -54.23 | <.0001 | 0.05 | -3.3594 | -3.0942 | -0.00009 |
| beta2 | 0.4806 | 0.1989 | 10 | 2.42 | 0.0363 | 0.05 | 0.03745 | 0.9238 | 3.645E-7 |
| beta3 | -2.4592 | 0.05126 | 10 | -47.97 | <.0001 | 0.05 | -2.5734 | -2.3449 | 0.000039 |
| s2b1 | 0.02803 | 0.01221 | 10 | 2.30 | 0.0445 | 0.05 | 0.000833 | 0.05523 | -0.00014 |
| cb12 | -0.00127 | 0.03404 | 10 | -0.04 | 0.9710 | 0.05 | -0.07712 | 0.07458 | -0.00007 |
| s2b2 | 0.4331 | 0.2005 | 10 | 2.16 | 0.0560 | 0.05 | -0.01353 | 0.8798 | -6.98E-6 |
| s2 | 0.5016 | 0.06837 | 10 | 7.34 | <.0001 | 0.05 | 0.3493 | 0.6540 | 6.133E-6 |
The "Parameter Estimates" table contains the maximum likelihood estimates of the parameters (Output 63.1.5). Both s2b1 and s2b2 are marginally significant, indicating between-subject variability in the clearances and absorption rate constants, respectively. There does not appear to be a significant covariance between them, as seen by the estimate of cb12.
The estimates of 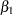 ,
, 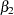 , and
, and 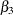 are close to the adaptive quadrature estimates listed in Table 3 of Pinheiro and Bates (1995). However, Pinheiro and Bates use a Cholesky-root parameterization for the random-effects variance matrix and a logarithmic parameterization for the residual variance. The PROC NLMIXED statements using their parameterization are as follows, and results are similar.
are close to the adaptive quadrature estimates listed in Table 3 of Pinheiro and Bates (1995). However, Pinheiro and Bates use a Cholesky-root parameterization for the random-effects variance matrix and a logarithmic parameterization for the residual variance. The PROC NLMIXED statements using their parameterization are as follows, and results are similar.
proc nlmixed data=theoph; parms ll1=-1.5 l2=0 ll3=-0.1 beta1=-3 beta2=0.5 beta3=-2.5 ls2=-0.7; s2 = exp(ls2); l1 = exp(ll1); l3 = exp(ll3); s2b1 = l1*l1*s2; cb12 = l2*l1*s2; s2b2 = (l2*l2 + l3*l3)*s2; cl = exp(beta1 + b1); ka = exp(beta2 + b2); ke = exp(beta3); pred = dose*ke*ka*(exp(-ke*time)-exp(-ka*time))/cl/(ka-ke); model conc ~ normal(pred,s2); random b1 b2 ~ normal([0,0],[s2b1,cb12,s2b2]) subject=subject; run;