The GAMPL Procedure
-
Overview

- Getting Started
-
Syntax

-
Details
 Missing ValuesThin-Plate Regression SplinesGeneralized Additive ModelsModel Evaluation CriteriaFitting AlgorithmsDegrees of FreedomModel InferenceDispersion ParameterTests for Smoothing ComponentsComputational Method: MultithreadingChoosing an Optimization TechniqueDisplayed OutputODS Table NamesODS Graphics
Missing ValuesThin-Plate Regression SplinesGeneralized Additive ModelsModel Evaluation CriteriaFitting AlgorithmsDegrees of FreedomModel InferenceDispersion ParameterTests for Smoothing ComponentsComputational Method: MultithreadingChoosing an Optimization TechniqueDisplayed OutputODS Table NamesODS Graphics -
Examples

- References
PROC GAMPL Statement
-
PROC GAMPL <options>;
The PROC GAMPL statement invokes the procedure. Table 7.1 summarizes the available options in the PROC GAMPL statement by function. The options are then described fully in alphabetical order.
Table 7.1: PROC GAMPL Statement Options
|
Option |
Description |
|---|---|
|
Basic Options |
|
|
Specifies a global significance level |
|
|
Specifies the input data set |
|
|
Limits the length of effect names |
|
|
Sets the seed for pseudorandom number generation |
|
|
Display Options |
|
|
Displays the "Iteration History" table |
|
|
Limits or suppresses the display of classification variable levels |
|
|
Suppresses ODS output |
|
|
Controls plots that are produced through ODS Graphics |
|
|
Optimization Subject Options |
|
|
Sets optimization parameters for likelihood estimation |
|
|
Sets optimization parameters for smoothing parameter estimation |
|
|
Tolerance Options |
|
|
Tunes the singularity criterion for Cholesky decompositions |
|
|
Tunes the singularity criterion for the sweep operator |
|
|
Tunes the general singularity criterion |
|
|
User-Defined Format Options |
|
|
Specifies the file reference for a format stream |
|
You can specify the following options in the PROC GAMPL statement.
Optimization Parameters
You can specify optimization-parameters for both the PLIKEOPTIONS and SMOOTHOPTIONS options. Depending on the modeling context, some optimization parameters might have no effect. For parametric generalized linear models or generalized additive models that have fixed smoothing parameters, any optimization parameters that you specify in the SMOOTHOPTIONS option are ignored. For the performance iteration method , only the ABSFCONV= , FCONV= , and MAXITER= options are effective for PLIKEOPTIONS . The optimization algorithm is considered to have converged when any one of the convergence criteria that are specified in optimization-parameters is satisfied. Table 7.2 lists the available optimization parameters for both the PLIKEOPTIONS and SMOOTHOPTIONS options.
Table 7.2: Optimization Parameters
|
Option |
Description |
|---|---|
|
Tunes the absolute function convergence criterion |
|
|
Tunes the absolute function difference convergence criterion |
|
|
Tunes the absolute gradient convergence criterion |
|
|
Tunes the relative function difference convergence criterion |
|
|
Tunes the relative gradient convergence criterion |
|
|
Specifies the maximum number of function evaluations in any optimization |
|
|
Chooses the maximum number of iterations in any optimization |
|
|
Specifies the upper limit of CPU time (in seconds) for any optimization |
|
|
Specifies the minimum number of iterations in any optimization |
|
|
Selects the optimization technique |
You can specify the following optimization-parameters:
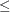
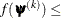
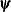
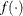
![\[ |f(\bpsi ^{(k-1)}) - f(\bpsi ^{(k)})| \leq r \]](images/stathpug_hpgam0006.png)
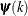
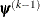
![\[ \max _ j |g_ j(\bpsi ^{(k)})| \leq r \]](images/stathpug_hpgam0009.png)
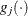
![\[ \frac{|f(\bpsi ^{(k)}) - f(\bpsi ^{(k-1)})|}{|f(\bpsi ^{(k-1)})|} \leq r \]](images/stathpug_hpgam0011.png)
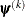
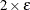

![\[ \frac{\mb{g}(\bpsi ^{(k)})^\prime [\bH ^{(k)}]^{-1} \mb{g}(\bpsi ^{(k)})}{|f(\bpsi ^{(k)})| } \leq r \]](images/stathpug_hpgam0015.png)
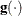
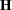
![\[ \frac{\parallel \mb{g}(\bpsi ^{(k)}) \parallel _2^2 \quad \parallel \mb{s}(\bpsi ^{(k)}) \parallel _2}{\parallel \mb{g}(\bpsi ^{(k)}) - \mb{g}(\bpsi ^{(k-1)}) \parallel _2 |f(\bpsi ^{(k)})| } \leq r \]](images/stathpug_hpgam0018.png)