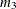-
HOLD<=value-list>
EQCONS<=value-list>
-
specifies which parameter values PROC HPMIXED
should hold equal to the specified values. If you do not specify value-list, then all covariance parameters are held equal to the specified values. For example, the following statement constrains the
first and third covariance parameters to equal 5 and 2, respectively:
parms (5) (3) (2) (3) / hold=1,3;
Specifying the HOLD= option implies the NOPROFILE
option in the PROC HPMIXED
statement.
-
LOWERB=value-list
-
enables you to specify lower boundary constraints on the covariance parameters. The value-list specification is a list of numbers or missing values (.) separated by commas. You must list the numbers in the order that
PROC HPMIXED uses for the covariance parameters, and each number corresponds to the lower boundary constraint. A missing value
instructs PROC HPMIXED to use its default constraint, and if you do not specify numbers for all the covariance parameters,
PROC MIXED assumes that the remaining ones are missing.
-
NOITER
-
requests that no optimization iterations be performed and that PROC HPMIXED use the best value from the grid search to perform
inferences. By default, iterations begin at the best value from the PARMS grid search. This option is ignored when you specify
the HOLD=
option.
If a residual variance is profiled, the parameter estimates can change from the initial values that you provide as the residual
variance is recomputed. To prevent an update of the residual variance, combine the NOITER option with the NOPROFILE
option in the PROC HPMIXED
statements, as in the following program:
proc hpmixed noprofile;
class A B C rep mp sp;
model y = A | B | C;
random rep mp sp;
parms (180) (200) (170) (1000) / noiter;
run;
Specifying the NOITER option in the PARMS statement has the same effect as specifying TECHNIQUE=NONE in the NLOPTIONS
statement.
Notice that the NOITER option can be useful if you want to obtain the starting values HPMIXED computes. The following statements
produce the starting values:
proc hpmixed noprofile;
class A B;
model y = A;
random int / subject=B;
parms / noiter;
run;
-
PARMSDATA=SAS-data-set
PDATA=SAS data set
-
reads in covariance parameter values from a SAS data set. The data set should contain the numerical variable ESTIMATE or the
numerical variables Covp1–Covpq, where q denotes the number of covariance parameters.
If the PARMSDATA= data set contains multiple sets of covariance parameters, the HPMIXED procedure evaluates the initial objective
function for each set and commences the optimization step by using the set with the lowest function value as the starting
values. For example, the following SAS statements request that the objective function be evaluated for three sets of initial
values:
data data_covp;
input covp1-covp4;
datalines;
180 200 170 1000
170 190 160 900
160 180 150 800
;
proc hpmixed;
class A B C rep;
model yield = A;
random rep B C;
parms / pdata=data_covp;
run;
Each set comprises four covariance parameters.
The order of the observations in a data set with the numerical variable Estimate corresponds to the order of the covariance parameters in the "Covariance Parameter Estimates" table.
The PARMSDATA= data set must contain at least one set of covariance parameters with no missing values.
If the HPMIXED procedure is processing the input data set in BY
groups, you can add the BY variables to the PARMSDATA= data set. If this data set is sorted by the BY variables, the HPMIXED
procedure matches the covariance parameter values to the current BY group. If the PARMSDATA= data set does not contain all
BY variables, the data set is processed in its entirety for every BY group and a message is written to the log. This enables
you to provide a single set of starting values across BY groups, as in the following statements:
data data_covp;
input covp1-covp4;
datalines;
180 200 170 1000
;
proc hpmixed;
class A B C rep;
model yield = A;
random rep B C;
parms / pdata=data_covp;
by year;
run;
The same set of starting values is used for each value of the year variable.
-
UPPERB=value-list
-
enables you to specify upper boundary constraints on the covariance parameters. The value-list specification is a list of numbers or missing values (.) separated by commas. You must list the numbers in the order that
PROC HPMIXED uses for the covariance parameters, and each number corresponds to the upper boundary constraint. A missing value
instructs PROC HPMIXED to use its default constraint, and if you do not specify numbers for all of the covariance parameters,
PROC HPMIXED assumes that the remaining ones are missing.
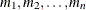
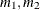 to
to