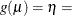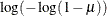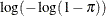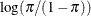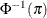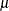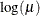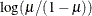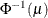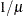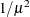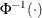The GEE Procedure
MODEL Statement
-
MODEL response = <effects> </ options>;
-
MODEL events/trials = <effects> </ options>;
The MODEL statement specifies the response (dependent variable) and the effects (explanatory variables). If you omit the explanatory variables, PROC GEE fits an intercept-only model. An intercept term is included in the model by default. You can remove the intercept by specifying the NOINT option.
You can specify the response in the form of a single variable (response) or in the form of a ratio of two variables ( events/trials). The first form is applicable to all responses. The second form is applicable only to summarized binomial response data. When each observation in the input data set contains the number of events (for example, successes) and the number of trials from a set of binomial trials, use the events/trials syntax.
In the events/trials model syntax, you specify two variables: one for the event counts and one for trial counts. These two variables are separated by a slash (/). The value of the events variable must be nonnegative, and the value of the trials variable must be equal to or greater than the value of the events variable for an observation to be valid. The events and trials variables can take non-integer values.
When each observation in the input data set contains a single trial from a binomial experiment, use the response form of the MODEL statement. The response variable can be numeric or character. The ordering of response levels is critical in these models.
Responses for the Poisson distribution must be all nonnegative, but they can be non-integer values.
The effects in the MODEL statement consist of an explanatory variable or combination of variables. Explanatory variables can be continuous or classification variables. Classification variables can be character or numeric. Explanatory variables that represent nominal (classification) data must be declared in a CLASS statement. Interactions between variables can also be included as effects. Columns of the design matrix are automatically generated for classification variables and interactions. The syntax for specifying effects is the same as for the GLM procedure. For more information, see the section Specification of Effects in Chapter 46: The GLM Procedure.
Table 43.5 summarizes the options available in the MODEL statement.
Table 43.5: MODEL Statement Options
|
Option |
Description |
|---|---|
|
Sets the confidence coefficient |
|
|
Specifies the probability distribution |
|
|
Specifies the link function |
|
|
Requests no intercept term |
|
|
Holds the scale parameter fixed |
|
|
Specifies a variable in the input data set to be used as an offset |
|
|
Specifies the value used for the scale |
You can specify the following options after a slash (/).