The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement
PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement -
Details
 Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsTweedie Distribution For Generalized Linear ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics
Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsTweedie Distribution For Generalized Linear ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics -
Examples
 Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson RegressionTweedie Regression
Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson RegressionTweedie Regression - References
Count data that have an incidence of zeros greater than expected for the underlying probability distribution of counts can
be modeled with a zero-inflated distribution. In GENMOD, the underlying distribution can be either Poisson or negative binomial.
See Lambert (1992), Long (1997) and Cameron and Trivedi (1998) for more information about zero-inflated models. The population is considered to consist of two types of individuals. The
first type gives Poisson or negative binomial distributed counts, which might contain zeros. The second type always gives
a zero count. Let ![]() be the underlying distribution mean and
be the underlying distribution mean and ![]() be the probability of an individual being of the second type. The parameter
be the probability of an individual being of the second type. The parameter ![]() is called here the zero-inflation probability, and is the probability of zero counts in excess of the frequency predicted by the underlying distribution. You can request
that the zero inflation probability be displayed in an output data set with the PZERO keyword. The probability distribution of a zero-inflated Poisson random variable Y is given by
is called here the zero-inflation probability, and is the probability of zero counts in excess of the frequency predicted by the underlying distribution. You can request
that the zero inflation probability be displayed in an output data set with the PZERO keyword. The probability distribution of a zero-inflated Poisson random variable Y is given by
and the probability distribution of a zero-inflated negative binomial random variable Y is given by
where k is the negative binomial dispersion parameter.
You can model the parameters ![]() and
and ![]() in GENMOD with the regression models:
in GENMOD with the regression models:
where h is one of the binary link functions: logit, probit, or complementary log-log. The link function h is the logit link by default, or the link function option specified in the ZEROMODEL statement. The link function g is the log link function by default, or the link function specified in the MODEL statement, for both the Poisson and the
negative binomial. The covariates ![]() for observation i are determined by the model specified in the ZEROMODEL statement, and the covariates
for observation i are determined by the model specified in the ZEROMODEL statement, and the covariates ![]() are determined by the model specified in the MODEL statement. The regression parameters
are determined by the model specified in the MODEL statement. The regression parameters ![]() and
and ![]() are estimated by maximum likelihood.
are estimated by maximum likelihood.
The mean and variance of Y for the zero-inflated Poisson are given by
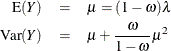
and for the zero-inflated negative binomial by
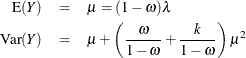
You can request that the mean of Y be displayed for each observation in an output data set with the PRED keyword.