SCALEMODEL regressor-variable-list </ scalemodel-option> ;
The SCALEMODEL statement specifies regression variables. All the variables specified in this statement must be present in
the input data set that is specified by the DATA= option in the PROC SEVERITY statement. The scale parameter of each candidate
distribution is linked to a linear combination of these regression variables along with an intercept. If a distribution does
not have a scale parameter, then a model based on that distribution is not estimated. If you specify more than one SCALEMODEL
statement, then the first statement is used.
The regressor variables are expected to have nonmissing values. If any of the variables has a missing value in an observation,
then a warning is written to the SAS log and that observation is ignored.
For more information about modeling regression effects, see the section Estimating Regression Effects.
You can specify the following scalemodel-option in the SCALEMODEL statement:
-
DFMIXTURE=method-name <(method-options)>
-
specifies the method for computing representative estimates of the cumulative distribution function (CDF) and the probability
density function (PDF).
When regression variables are specified, the scale of the distribution depends on the values of the regressors. For a given
distribution family, each observation in the input data set implies a different scaled version of the distribution. To compute
estimates of CDF and PDF that are comparable across different distribution families, PROC SEVERITY needs to construct a single
representative distribution from all such distributions. You can specify one of the following method-name values to specify the method that is used to construct the representative distribution. For more information about each of
the methods, see the section CDF and PDF Estimates with Regression Effects.
-
FULL
-
specifies that the representative distribution be the mixture of 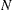 distributions such that each distribution has a scale value that is implied by each of the
distributions such that each distribution has a scale value that is implied by each of the 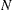 observations that are used for estimation. This method is the slowest.
observations that are used for estimation. This method is the slowest.
-
MEAN
-
specifies that the representative distribution be the one-point mixture of the distribution whose scale value is the mean
of the 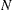 scale values that are implied by the
scale values that are implied by the 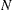 observations that are used for estimation. If you do not specify the DFMIXTURE= option, then this method is used by default.
This is also the fastest method.
observations that are used for estimation. If you do not specify the DFMIXTURE= option, then this method is used by default.
This is also the fastest method.
-
QUANTILE <(K=q)>
-
specifies that the representative distribution be the mixture of a fixed number of distributions whose scale values are the
quantiles from the sample of 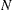 scale values that are implied by the
scale values that are implied by the 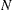 observations in the current BY group (or in the entire DATA= data set if the BY statement is not specified).
observations in the current BY group (or in the entire DATA= data set if the BY statement is not specified).
You can use the K= option to specify the number of distributions in the mixture. If you specify K=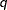 , then the mixture contains
, then the mixture contains 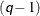 distributions such that each distribution has as its scale one of the
distributions such that each distribution has as its scale one of the 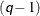 -quantiles.
-quantiles.
If you do not specify the K= option, then PROC SEVERITY uses the default of 2, which implies the use of a one-point mixture
with a distribution whose scale value is the median of all scale values.
-
RANDOM <(random-method-options)>
-
specifies that the representative distribution be the mixture of a fixed number of distributions whose scale values are the
scale values that are implied by a randomly chosen subset of the set of all observations in the current BY group (or in the
entire DATA= data set if the BY statement is not specified). The same subset of observations is used for each distribution
family.
You can specify the following random-method-options to specify how the subset is chosen:
-
K=r
-
specifies the number of distributions to include in the mixture. If you do not specify this option, then PROC SEVERITY uses
the default of 15.
-
SEED=number
-
specifies the seed that is used to generate the uniform random sample of observation indices. If you do not specify this option,
then PROC SEVERITY generates a seed internally that is based on the current value of the system clock.
Copyright © SAS Institute Inc. All Rights Reserved.


 Predefined DistributionsCensoring and TruncationParameter Estimation MethodParameter InitializationEstimating Regression EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDefining a Distribution Model with the FCMP ProcedurePredefined Utility FunctionsCustom Objective FunctionsMultithreaded ComputationInput Data SetsOutput Data SetsDisplayed OutputODS Graphics
Predefined DistributionsCensoring and TruncationParameter Estimation MethodParameter InitializationEstimating Regression EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDefining a Distribution Model with the FCMP ProcedurePredefined Utility FunctionsCustom Objective FunctionsMultithreaded ComputationInput Data SetsOutput Data SetsDisplayed OutputODS Graphics