The HTSNP Procedure (Experimental)
PROC HTSNP <options> ;
You can specify the following options in the PROC HTSNP statement.
- BEST=number
-
specifies the number of the best selections displayed in the “htSNP Evaluation” table during an exhaustive or simulated annealing search process when METHOD=EX or SA, respectively, is specified. The number must be a positive integer. By default, only one best selection is reported. Note that sets of SNPs with the same value of the criterion measure as the last displayed set(s) are not necessarily all shown since number indicates the number of sets actually displayed. If number is greater than the number of possible sets when METHOD=EX or greater than the number of sets examined when METHOD=SA, there are fewer than number sets displayed.
- CONV=number
-
specifies the convergence criterion for search of htSNPs, where
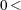 number
number 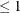 . The search process is stopped when the haplotype criterion is greater than or equal to number specified in the CONV= option. The default value is 0.90. When METHOD=SA or METHOD=EX is specified, the CONV= option is ignored
and the searching continues until the annealing schedule is finished or the whole search space is traversed.
. The search process is stopped when the haplotype criterion is greater than or equal to number specified in the CONV= option. The default value is 0.90. When METHOD=SA or METHOD=EX is specified, the CONV= option is ignored
and the searching continues until the annealing schedule is finished or the whole search space is traversed.
-
CRITERION=PDE | RSQH
CRIT=PDE | RSQH -
indicates the criterion to use for evaluating candidate sets of htSNPs. By default or when CRITERION=PDE is specified, the proportion of diversity explained (PDE) is used (Clayton, 2002). When CRITERION=RSQH, Stram et al.’s
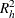 is used to measure haplotype richness (2003). See the section Evaluating Sets of htSNPs for more information about these measures.
is used to measure haplotype richness (2003). See the section Evaluating Sets of htSNPs for more information about these measures.
- CUTOFF=number
-
specifies a lower bound on a haplotype’s frequency in order for that haplotype to be included in the search process for sets of htSNPs. The value of number must be between 0 and 1. By default, all haplotypes from the sample are included in the search process.
- DATA=SAS-data-set
-
names the input SAS data set to be used by PROC HTSNP. The default is to use the most recently created data set.
- MAXSIZE=number
-
specifies the maximum number of markers to be included in the subset for incremental search by default or when METHOD=INCR is specified. The number must be a positive integer that is less than or equal to the number of markers specified in the VAR statement. Searching is carried out until convergence is reached according to the convergence criterion, or until number of markers have been included in the subset.
-
METHOD=INCR | INCREMENTAL
METHOD=DECR | DECREMENTAL
METHOD=EX | EXHAUSTIVE
METHOD=IM | ITERMAX
METHOD=SA | SIMANNEAL -
indicates the method used for core marker set selection. By default or when METHOD=INCR is specified, the incremental search algorithm is used. When METHOD=DECR, the decremental algorithm is used. When METHOD=EX, the exhaustive search algorithm is used. When METHOD=IM, the iterative maximization algorithm is used. When METHOD=SA, the simulated annealing search algorithm is used. See the section Search Algorithms for more information about these methods.
-
NOSUMMARY
NOSUMM - SCHEDULE=number
-
specifies the number of reconfigurations used in each annealing step when METHOD=SA. The value for number must be a positive integer. The default value is 100
 (number of variables specified in the VAR statement).
(number of variables specified in the VAR statement).
- SEED=number
-
specifies the initial seed for the random number generator used for the sampling of markers. The value for number must be an integer; the computer clock time is used if the option is omitted or an integer less than or equal to 0 is specified. For more details about seed values, see SAS Language Reference: Concepts.
- SIZE=number
-
specifies the size of the subset of markers to select. The value for number must be a positive integer that is less than or equal to the number of markers specified in the VAR statement. The SIZE= option must be specified for an exhaustive search, iterative maximization search, and simulated annealing search.
- STEP=number
-
specifies the steps used for simulated annealing search when METHOD=SA. The value for number must be a positive integer. The default value is 1.
-
TEMPERATURE=number
T=number -
specifies the temperature used for the simulated annealing search when METHOD=SA is specified. The value for number must be a positive number. The default value is 1.
- TFACTOR=number
-
specifies the factor by which the temperature is reduced for each annealing step during simulated annealing search when METHOD=SA. The value for number must satisfy
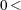 number
number 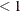 . The default value is 0.90.
. The default value is 0.90.
