The SURVEYIMPUTE Procedure
Fully Efficient Fractional Imputation
The fully efficient fractional imputation (FEFI) method uses multiple donor units for a recipient unit. The observation unit that contains the missing values is known as the recipient unit, and the observation unit that provides the value for imputation is known as the donor unit. The number of donor units for a recipient unit is equal to the number of observed levels for the missing items, given the observed levels for the nonmissing items of the recipient unit. Each donor donates a fraction of the original weight of the recipient unit such that the sum of the fractional weights from all the donors is equal to the original weight of the recipient. The fraction of the recipient weight that a donor unit contributes to the recipient unit is known as the fractional weight. The method is called fully efficient because it does not introduce additional variability that is caused by the selection of donor units (Kim and Fuller 2004). One disadvantage of the FEFI method is that it can greatly increase the size of the imputed data set. For more information, see Kalton and Kish (1984), Fuller (2009, Section 5.2.2), and Kim and Shao (2014, Section 4.6).
FEFI Algorithm
Suppose you want to impute P items jointly (by using the IMPJOINT
statement in PROC SURVEYIMPUTE). Let 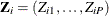 be the true response for the P items for unit i.
be the true response for the P items for unit i. 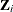 is completely known if all P items are observed for unit i. However, the true response might not be known for some units because of item nonresponse. Let
is completely known if all P items are observed for unit i. However, the true response might not be known for some units because of item nonresponse. Let 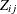 be categorical and have J levels for item j. Denote
be categorical and have J levels for item j. Denote 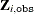 as the observed part and
as the observed part and 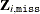 as the missing part of
as the missing part of 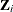 . Let the population proportion that falls in category
. Let the population proportion that falls in category 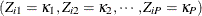 be
be 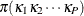 . Assume that it is possible to estimate the population proportion from the observed sample. That is, for example, the conditional
probability,
. Assume that it is possible to estimate the population proportion from the observed sample. That is, for example, the conditional
probability, 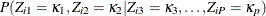 , in the observed data is the same as the conditional probability in the data where
, in the observed data is the same as the conditional probability in the data where 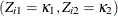 are missing. The conditional probabilities are estimated by
are missing. The conditional probabilities are estimated by
![\[ \hat P(Z_{i1}=\kappa _1,Z_{i2}=\kappa _2|Z_{i3}=\kappa _3,\ldots ,Z_{iP}=\kappa _ p) = \left\{ \sum _{\kappa _1 \kappa _2} \hat\pi (\kappa _1\kappa _2 \cdots \kappa _ P) \right\} ^{-1} \hat\pi (\kappa _1\kappa _2 \cdots \kappa _ P) \]](images/statug_surveyimpute0017.png)
where
![\[ \hat\pi (\kappa _1\kappa _2 \cdots \kappa _ P) = \left\{ \sum _ i w_ i \right\} ^{-1} \sum _ i w_ i I(Z_{i1}=\kappa _1,\ldots ,Z_{iP}=\kappa _ P) \]](images/statug_surveyimpute0018.png)
is the estimated joint probability, 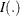 is an indicator function, and
is an indicator function, and 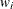 is the observation weight for unit i.
is the observation weight for unit i.
The FEFI method uses an EM-by-weighting algorithm similar to that of Ibrahim (1990). The detailed algorithm is described in Kim and Fuller (2013). The following steps describe the imputation technique. If you do not specify imputation cells by using the CELLS statement, PROC SURVEYIMPUTE uses the entire data set as one imputation cell. If you specify imputation cells, then all the probabilities in these steps are computed by using observations from the same imputation cell as the recipient unit. To simplify notation, subscripts are not used for imputation cells in the following description.
For given i, let 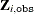 and
and 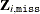 be the observed part and the missing part, respectively, of unit i. Let
be the observed part and the missing part, respectively, of unit i. Let 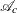 be the index set for the complete respondents. Suppose you want to impute the missing part of
be the index set for the complete respondents. Suppose you want to impute the missing part of 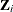 ,
, 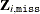 . The index set
. The index set 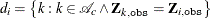 contains the indexes for the all possible donor units for
contains the indexes for the all possible donor units for 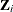 . Let
. Let 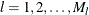 be all the observed combinations of
be all the observed combinations of 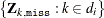 . The set of all observed combinations for unit i defines the donor cells (all possible realizations) for unit i. Let
. The set of all observed combinations for unit i defines the donor cells (all possible realizations) for unit i. Let ![$\bZ _{i,\mt {miss}[l]}$](images/statug_surveyimpute0025.png) be the lth imputed value of
be the lth imputed value of 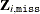 . You must assume that at least one imputed value is available; otherwise the observation is not imputed.
. You must assume that at least one imputed value is available; otherwise the observation is not imputed.
-
Initialization: For each observation that has missing items, determine the number of donor cells by using the number of unique combinations of observed levels for the missing items for the responding units in the imputation cell. Compute the initial fractional weight from donor cell l to unit i,
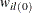 , by
, by
![\[ w_{il(0)} = \left\{ \sum _{k=1}^{M_ l} \tilde\pi _{(0)} (\bZ _{i,\mt {obs}},\bZ _{i,\mt {miss}[k]}) \right\} ^{-1} \tilde\pi _{(0)} (\bZ _{i,\mt {obs}},\bZ _{i,\mt {miss}[l]}) \]](images/statug_surveyimpute0027.png)
where
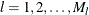 is the number of donor cells, and
is the number of donor cells, and
![\[ \tilde\pi _{(0)} (\kappa _1,\ldots ,\kappa _ P) = \left\{ \sum _{i \in \mathcal{A}_ c} w_ i \right\} ^{-1} \sum _{i \in \mathcal{A}_ c} w_ i I(Z_{i1}=\kappa _1,\ldots ,Z_{iP}=\kappa _ P) \]](images/statug_surveyimpute0028.png)
The sum of the fractional weights over all the donor cells is 1 for every observation unit; that is,
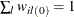 , for all i. The lth imputed row for unit i is created by keeping the observed items unchanged, replacing the missing items with the observed levels from the lth donor cell, and computing the fractional weight by
, for all i. The lth imputed row for unit i is created by keeping the observed items unchanged, replacing the missing items with the observed levels from the lth donor cell, and computing the fractional weight by 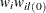 . Only the complete respondents are used to compute the fractional weights in this step. If unit i has no missing items, then
. Only the complete respondents are used to compute the fractional weights in this step. If unit i has no missing items, then 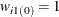 . The initial FEFI data set contains all the observed units, the imputed rows for observation that had missing items, and
the corresponding fractional weights.
. The initial FEFI data set contains all the observed units, the imputed rows for observation that had missing items, and
the corresponding fractional weights.
-
M-step: The tth M-step computes the joint probabilities by using the fractional weights from the (t–1)th E-step,
![\[ \tilde\pi _{(t)} (\kappa _1,\ldots ,\kappa _ P) = \left\{ \sum _ i \sum _ l w_ i w_{il(t-1)} \right\} ^{-1} \sum _ i \sum _ l w_ i w_{il(t-1)} I(Z_{i1}=\kappa _1,\ldots ,Z_{iP}=\kappa _ P) \]](images/statug_surveyimpute0032.png)
for all i, all l, and
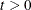 . Note that for
. Note that for 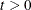 ,
, 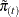 uses all observation units including observations that have missing items that are imputed in the initialization step.
uses all observation units including observations that have missing items that are imputed in the initialization step.
-
E-step: The tth E-step computes the fractional weights by using the joint probabilities
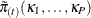 from the tth M-step. The tth fractional weight for unit i and donor cell l is given by
from the tth M-step. The tth fractional weight for unit i and donor cell l is given by
![\[ w_{il(t)} = \left\{ \sum _{k=1}^{M_ l} \tilde\pi _{(t)} (\bZ _{i,\mt {obs}},\bZ _{i,\mt {miss}[k]}) \right\} ^{-1} \tilde\pi _{(t)} (\bZ _{i,\mt {obs}},\bZ _{i,\mt {miss}[l]}) \]](images/statug_surveyimpute0036.png)
-
Repetition: The EM steps are repeated for
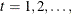 until the changes in fractional weights over all observation units between two successive EM steps are negligible or the
maximum number of EM repetitions is reached.
until the changes in fractional weights over all observation units between two successive EM steps are negligible or the
maximum number of EM repetitions is reached.
The maximum absolute difference convergence criterion,
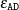 , at step t is defined as
, at step t is defined as
![\[ \max _{i,l} \lvert {w_{il(t)}-w_{il(t-1)}} \rvert / {w_{il}} \leq \epsilon _{\mt {AD}} \]](images/statug_surveyimpute0039.png)
The maximum absolute relative difference convergence criterion,
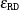 , at step t is defined as
, at step t is defined as
![\[ \max _{i,l} \lvert {w_{il(t)}-w_{il(t-1)}} \rvert /{w_{il(t-1)}} \leq \epsilon _{\mt {RD}} \]](images/statug_surveyimpute0041.png)
where
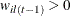 .
.
The replicate weights are created by computing a replicated version of 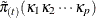 ,
, 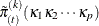 , and by repeating the EM-by-weighting algorithm as described earlier. For the kth replicate sample,
, and by repeating the EM-by-weighting algorithm as described earlier. For the kth replicate sample, 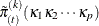 is computed by
is computed by
![\[ \tilde\pi _{(t)}^{(k)} (\kappa _1,\ldots ,\kappa _ P) = \left\{ \sum _ i \sum _ l w_ i^{(k)} w_{il(t-1)}^{(k)} \right\} ^{-1} \sum _ i \sum _ l w_ i^{(k)} w_{il(t-1)}^{(k)} I(Z_{i1}=\kappa _1,\ldots ,Z_{iP}=\kappa _ P) \]](images/statug_surveyimpute0045.png)
Example of FEFI
The small data set shown in Figure 110.8 is used to illustrate the imputation technique. The data set contains nine observation units, and each unit has two items
(X and Y). The variable Unit contains the observation identification. In this example, X is missing for units 5 and 9, and Y is missing for units 2 and 9.
Figure 110.8: Sample Data with Missing Items
The following SAS statements request joint imputation of X and Y by using the FEFI method. These statements also request imputation-adjusted replicate weights for the jackknife replication
method. The CLASS statement specifies that both X and Y are CLASS variables. The OUTPUT
statement stores the imputed values in the data set Imputed and stores the jackknife coefficients in the data set Ojkc. The FRACTIONALWEIGHTS= option in the OUTPUT
statement saves the fractional weights in the Imputed data set.
proc surveyimpute data=test varmethod=jackknife; class x y; var x y; id Unit; output out=Imputed fractionalweights=FracWgt outjkcoefs=Ojkc; run;
The initial fractional weights, FracWgt, after the initialization step are displayed in Figure 110.9.
-
Observation unit 1 has no missing value. Therefore, the
Recipientvalue is 0, theFracWgtvalue is 1, and the values ofXandYare the same as the observed values for observation unit 1 in Figure 110.9. Because all observation units have a weight of 1, the fractional weights,FracWgt, and the imputation-adjusted weights,ImpWt, are the same for all rows. -
Observation unit 2 has a missing
Y. The observed level forXfor unit 2 is 0. ForX= 0, two levels forYare observed:Y= 0, which has a proportion (FracWgt) of 0.67, andY= 1, which has a proportion of 0.33. Therefore, observation unit 2 receives two donor cells (Recipient= 1 andRecipient= 2), whose initial fractional weights are 0.67 and 0.33, respectively. BecauseXis observed, theXvalues in both rows for unit 2 are the same as the observed value. However, the first recipient row for unit 2 has an imputedYvalue of 0, the second recipient row for unit 2 has an imputedYvalue of 1, and each has a corresponding initial fractional weight. -
Observation unit 5 has a missing
X. The observed level forYfor unit 5 is 1. To imputeX, note that two levels ofXare observed whenY= 1:X= 0 with a proportion of 0.33 andX= 1 with a proportion of 0.67. The two recipient rows for observation unit 5 contain the initial fractional weights in theFracWgtcolumn and the imputedXvalues. -
Observation unit 9 has missing values for both
XandY. From the observed data,XandYcan take the following values: (X= 0,Y= 0) with probability 0.33, (X= 0,Y= 1) with probability 0.17, (X= 1,Y= 0) with probability 0.17, and (X= 1,Y= 1) with probability 0.33. The four imputed rows (Recipient1,Recipient2,Recipient3, andRecipient4) for observation unit 9 represent the four observed combinations forXandYalong with their initial fractional weights.
The resulting data set contains 14 rows. There are six rows for fully observed units (Recipient = 0), two rows for unit 2, two rows for unit 5, and four rows for unit 9. The sum of initial fractional weights is 1 for
all units.
Figure 110.9: Fractional Imputation after Initialization
| Unit | Recipient | ImpWt | FracWgt | X | Y |
|---|---|---|---|---|---|
| 1 | 0 | 1.00000 | 1.00000 | 0 | 0 |
| 2 | 1 | 0.66667 | 0.66667 | 0 | 0 |
| 2 | 2 | 0.33333 | 0.33333 | 0 | 1 |
| 3 | 0 | 1.00000 | 1.00000 | 0 | 1 |
| 4 | 0 | 1.00000 | 1.00000 | 0 | 0 |
| 5 | 1 | 0.33333 | 0.33333 | 0 | 1 |
| 5 | 2 | 0.66667 | 0.66667 | 1 | 1 |
| 6 | 0 | 1.00000 | 1.00000 | 1 | 0 |
| 7 | 0 | 1.00000 | 1.00000 | 1 | 1 |
| 8 | 0 | 1.00000 | 1.00000 | 1 | 1 |
| 9 | 1 | 0.33333 | 0.33333 | 0 | 0 |
| 9 | 2 | 0.16667 | 0.16667 | 0 | 1 |
| 9 | 3 | 0.16667 | 0.16667 | 1 | 0 |
| 9 | 4 | 0.33333 | 0.33333 | 1 | 1 |
The EM algorithm repeats the computation of the joint probabilities and the fractional weights until convergence. The fractional
weights, FracWgt, after the EM step and the imputation-adjusted replicate weights (ImpRepWt_1, …, ImpRepWt_9) are displayed in Figure 110.10.
Figure 110.10: Fractional Imputation after the EM
| Unit | Recipient | ImpWt | FracWgt | X | Y | ImpRepWt_1 | ImpRepWt_2 | ImpRepWt_3 | ImpRepWt_4 | ImpRepWt_5 | ImpRepWt_6 | ImpRepWt_7 | ImpRepWt_8 | ImpRepWt_9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 1.00000 | 1.00000 | 0 | 0 | 0.00000 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 |
| 2 | 1 | 0.58601 | 0.58601 | 0 | 0 | 0.46072 | 0.00000 | 1.12498 | 0.46072 | 0.75009 | 0.65906 | 0.62682 | 0.62682 | 0.65877 |
| 2 | 2 | 0.41399 | 0.41399 | 0 | 1 | 0.66428 | 0.00000 | 0.00002 | 0.66428 | 0.37491 | 0.46594 | 0.49818 | 0.49818 | 0.46623 |
| 3 | 0 | 1.00000 | 1.00000 | 0 | 1 | 1.12500 | 1.12500 | 0.00000 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 |
| 4 | 0 | 1.00000 | 1.00000 | 0 | 0 | 1.12500 | 1.12500 | 1.12500 | 0.00000 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 |
| 5 | 1 | 0.41399 | 0.41399 | 0 | 1 | 0.49821 | 0.37510 | 0.00002 | 0.49821 | 0.00000 | 0.46601 | 0.66443 | 0.66443 | 0.46623 |
| 5 | 2 | 0.58601 | 0.58601 | 1 | 1 | 0.62679 | 0.74990 | 1.12498 | 0.62679 | 0.00000 | 0.65899 | 0.46057 | 0.46057 | 0.65877 |
| 6 | 0 | 1.00000 | 1.00000 | 1 | 0 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 0.00000 | 1.12500 | 1.12500 | 1.12500 |
| 7 | 0 | 1.00000 | 1.00000 | 1 | 1 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 0.00000 | 1.12500 | 1.12500 |
| 8 | 0 | 1.00000 | 1.00000 | 1 | 1 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 1.12500 | 0.00000 | 1.12500 |
| 9 | 1 | 0.32330 | 0.32330 | 0 | 0 | 0.22659 | 0.32143 | 0.48214 | 0.22659 | 0.42862 | 0.41563 | 0.41109 | 0.41109 | 0.00000 |
| 9 | 2 | 0.22840 | 0.22840 | 0 | 1 | 0.32669 | 0.21434 | 0.00001 | 0.32669 | 0.21424 | 0.29384 | 0.32672 | 0.32672 | 0.00000 |
| 9 | 3 | 0.12500 | 0.12500 | 1 | 0 | 0.16071 | 0.16071 | 0.16071 | 0.16071 | 0.16071 | 0.00000 | 0.16071 | 0.16071 | 0.00000 |
| 9 | 4 | 0.32330 | 0.32330 | 1 | 1 | 0.41101 | 0.42851 | 0.48214 | 0.41101 | 0.32143 | 0.41553 | 0.22648 | 0.22648 | 0.00000 |
