The LOGISTIC Procedure
- Overview
- Getting Started
-
Syntax
 PROC LOGISTIC StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementEFFECT StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementODDSRATIO StatementOUTPUT StatementROC StatementROCCONTRAST StatementSCORE StatementSLICE StatementSTORE StatementSTRATA StatementTEST StatementUNITS StatementWEIGHT Statement
PROC LOGISTIC StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementEFFECT StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementNLOPTIONS StatementODDSRATIO StatementOUTPUT StatementROC StatementROCCONTRAST StatementSCORE StatementSLICE StatementSTORE StatementSTRATA StatementTEST StatementUNITS StatementWEIGHT Statement -
Details
 Missing ValuesResponse Level OrderingLink Functions and the Corresponding DistributionsDetermining Observations for Likelihood ContributionsIterative Algorithms for Model FittingConvergence CriteriaExistence of Maximum Likelihood EstimatesEffect-Selection MethodsModel Fitting InformationGeneralized Coefficient of DeterminationScore Statistics and TestsConfidence Intervals for ParametersOdds Ratio EstimationRank Correlation of Observed Responses and Predicted ProbabilitiesLinear Predictor, Predicted Probability, and Confidence LimitsClassification TableOverdispersionThe Hosmer-Lemeshow Goodness-of-Fit TestReceiver Operating Characteristic CurvesTesting Linear Hypotheses about the Regression CoefficientsJoint Tests and Type 3 TestsRegression DiagnosticsScoring Data SetsConditional Logistic RegressionExact Conditional Logistic RegressionInput and Output Data SetsComputational ResourcesDisplayed OutputODS Table NamesODS Graphics
Missing ValuesResponse Level OrderingLink Functions and the Corresponding DistributionsDetermining Observations for Likelihood ContributionsIterative Algorithms for Model FittingConvergence CriteriaExistence of Maximum Likelihood EstimatesEffect-Selection MethodsModel Fitting InformationGeneralized Coefficient of DeterminationScore Statistics and TestsConfidence Intervals for ParametersOdds Ratio EstimationRank Correlation of Observed Responses and Predicted ProbabilitiesLinear Predictor, Predicted Probability, and Confidence LimitsClassification TableOverdispersionThe Hosmer-Lemeshow Goodness-of-Fit TestReceiver Operating Characteristic CurvesTesting Linear Hypotheses about the Regression CoefficientsJoint Tests and Type 3 TestsRegression DiagnosticsScoring Data SetsConditional Logistic RegressionExact Conditional Logistic RegressionInput and Output Data SetsComputational ResourcesDisplayed OutputODS Table NamesODS Graphics -
Examples
 Stepwise Logistic Regression and Predicted ValuesLogistic Modeling with Categorical PredictorsOrdinal Logistic RegressionNominal Response Data: Generalized Logits ModelStratified SamplingLogistic Regression DiagnosticsROC Curve, Customized Odds Ratios, Goodness-of-Fit Statistics, R-Square, and Confidence LimitsComparing Receiver Operating Characteristic CurvesGoodness-of-Fit Tests and SubpopulationsOverdispersionConditional Logistic Regression for Matched Pairs DataExact Conditional Logistic RegressionFirth’s Penalized Likelihood Compared with Other ApproachesComplementary Log-Log Model for Infection RatesComplementary Log-Log Model for Interval-Censored Survival TimesScoring Data SetsUsing the LSMEANS StatementPartial Proportional Odds Model
Stepwise Logistic Regression and Predicted ValuesLogistic Modeling with Categorical PredictorsOrdinal Logistic RegressionNominal Response Data: Generalized Logits ModelStratified SamplingLogistic Regression DiagnosticsROC Curve, Customized Odds Ratios, Goodness-of-Fit Statistics, R-Square, and Confidence LimitsComparing Receiver Operating Characteristic CurvesGoodness-of-Fit Tests and SubpopulationsOverdispersionConditional Logistic Regression for Matched Pairs DataExact Conditional Logistic RegressionFirth’s Penalized Likelihood Compared with Other ApproachesComplementary Log-Log Model for Infection RatesComplementary Log-Log Model for Interval-Censored Survival TimesScoring Data SetsUsing the LSMEANS StatementPartial Proportional Odds Model - References
If you use the NOPRINT option in the PROC LOGISTIC statement, the procedure does not display any output. Otherwise, the tables displayed by the LOGISTIC procedure are discussed in the following section in the order in which they appear in the output. Some of the tables appear only in conjunction with certain options or statements; for more information, see the section ODS Table Names.
Note: The EFFECT, ESTIMATE, LSMEANS, LSMESTIMATE, and SLICE statements also create tables, which are not listed in this section. For information about these tables, see the corresponding sections of Chapter 19: Shared Concepts and Topics.
See the section Missing Values for information about missing-value handling, and the sections FREQ Statement and WEIGHT Statement for information about valid frequencies and weights.
Displays the Ordered Value assigned to each response level. For more information, see the section Response Level Ordering.
Displays the design values for each CLASS explanatory variable. For more information, see the section Other Parameterizations in Chapter 19: Shared Concepts and Topics.
The following tables are displayed if you specify the SIMPLE option in the PROC LOGISTIC statement:
-
Descriptive Statistics for Continuous Explanatory Variables
-
Frequency Distribution of Class Variables
-
Weight Distribution of Class Variables
Displays if you also specify a WEIGHT statement.
The following tables are displayed if you specify a STRATA statement:
-
Strata Summary
Shows the pattern of the number of events and the number of nonevents in a stratum. See the section STRATA Statement for more information. -
Strata Information
Displays if you specify the INFO option in a STRATA statement.
Displays if you specify the ITPRINT option in the MODEL statement. For more information, see the sections Iterative Algorithms for Model Fitting, Convergence Criteria, and Existence of Maximum Likelihood Estimates.
Displays if you specify the SCALE= option in the MODEL statement. Small p-values reject the null hypothesis that the fitted model is adequate. For more information, see the section Overdispersion.
Tests the parallel lines assumption if you fit an ordinal response model with the LINK= CLOGLOG or LINK= PROBIT options. If you specify LINK= LOGIT, this is called the "Proportional Odds" assumption. The table is not displayed if you specify the EQUALSLOPES or UNEQUALSLOPES option in the MODEL statement. Small p-values reject the null hypothesis that the slope parameters for each explanatory variable are constant across all the response functions. For more information, see the section Testing the Parallel Lines Assumption.
Computes various fit criteria based on a model with intercepts only and a model with intercepts and explanatory variables. If you specify the NOINT option in the MODEL statement, these statistics are calculated without considering the intercept parameters. For more information, see the section Model Fitting Information.
Tests the joint effect of the explanatory variables included in the model. Small p-values reject the null hypothesis that all slope parameters are equal to zero, ![]() . For more information, see the sections Model Fitting Information, Residual Chi-Square, and Testing Linear Hypotheses about the Regression Coefficients. If you also specify the RSQUARE
option in the MODEL
statement, two generalized R Square measures are included; for more information, see the section Generalized Coefficient of Determination.
. For more information, see the sections Model Fitting Information, Residual Chi-Square, and Testing Linear Hypotheses about the Regression Coefficients. If you also specify the RSQUARE
option in the MODEL
statement, two generalized R Square measures are included; for more information, see the section Generalized Coefficient of Determination.
Displays instead of the "Testing Global Null Hypothesis: BETA=0" table if the NOFIT
option is specified in the MODEL
statement. The global score test evaluates the joint significance of the effects in the MODEL statement. Small p-values reject the null hypothesis that all slope parameters are equal to zero, ![]() . For more information, see the section Residual Chi-Square.
. For more information, see the section Residual Chi-Square.
The tables in this section are produced when the SELECTION= option is specified in the MODEL statement. See the section Effect-Selection Methods for more information.
-
Residual Chi-Square Test
Displays if you specify SELECTION= FORWARD, BACKWARD, or STEPWISE in the MODEL statement. Small p-values reject the null hypothesis that the reduced model is adequate. For more information, see the section Residual Chi-Square. -
Analysis of Effects Eligible for Entry
Displays if you specify the DETAILS option and the SELECTION= FORWARD or STEPWISE option in the MODEL statement. Small p-values reject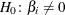 . The score chi-square is used to determine entry; for more information, see the section Testing Individual Effects Not in the Model.
. The score chi-square is used to determine entry; for more information, see the section Testing Individual Effects Not in the Model.
-
Analysis of Effects Eligible for Removal
Displays if you specify the SELECTION= BACKWARD or STEPWISE option in the MODEL statement. Small p-values reject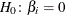 . The Wald chi-square is used to determine removal; for more information, see the section Testing Linear Hypotheses about the Regression Coefficients.
. The Wald chi-square is used to determine removal; for more information, see the section Testing Linear Hypotheses about the Regression Coefficients.
-
Analysis of Effects Removed by Fast Backward Elimination
Displays if you specify the FAST option and the SELECTION= BACKWARD or STEPWISE option in the MODEL statement. This table gives the approximate chi-square statistic for the variable removed, the corresponding p-value with respect to a chi-square distribution with one degree of freedom, the residual chi-square statistic for testing the joint significance of the variable and the preceding ones, the degrees of freedom, and the p-value of the residual chi-square with respect to a chi-square distribution with the corresponding degrees of freedom. -
Summary of Forward, Backward, and Stepwise Selection
Displays if you specify SELECTION= FORWARD, BACKWARD, or STEPWISE in the MODEL statement. The score chi-square is used to determine entry; for more information, see the section Testing Individual Effects Not in the Model. The Wald chi-square is used to determine removal; for more information, see the section Testing Linear Hypotheses about the Regression Coefficients. -
Regression Models Selected by Score Criterion
Displays the score chi-square for all models if you specify the SELECTION= SCORE option in the MODEL statement. Small p-values reject the null hypothesis that the fitted model is adequate. For more information, see the section Effect-Selection Methods.
Displays if the model contains a CLASS variable and performs Wald chi-square tests of the joint effect of the parameters for
each CLASS variable in the model. Small p-values reject ![]() . The title of this table for main-effects models or models that use GLM parameterization is "Type 3 Analysis of Effects";
for all other models the title is "Joint Tests." For more information, see the sections Testing Linear Hypotheses about the Regression Coefficients and Joint Tests and Type 3 Tests.
. The title of this table for main-effects models or models that use GLM parameterization is "Type 3 Analysis of Effects";
for all other models the title is "Joint Tests." For more information, see the sections Testing Linear Hypotheses about the Regression Coefficients and Joint Tests and Type 3 Tests.
CLASS effects are identified by their (nonreference) level. For generalized logit models, a response variable column displays the nonreference level of the logit. The table includes the following:
-
the estimated standard error of the parameter estimate, computed as the square root of the corresponding diagonal element of the estimated covariance matrix
-
the Wald chi-square statistic, computed by squaring the ratio of the parameter estimate divided by its standard error estimate. For more information, see the section Testing Linear Hypotheses about the Regression Coefficients.
-
the p-value tests the null hypothesis
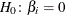 ; small values reject the null.
; small values reject the null.
-
the standardized estimate for the slope parameter, if you specify the STB option in the MODEL statement. For more information, see the STB option .
-
exponentiated values of the estimates of the slope parameters, if you specify the EXPB option in the MODEL statement. For more information, see the EXPB option .
-
the label of the variable, if you specify the PARMLABEL option in the MODEL statement and if space permits. Due to constraints on the line size, the variable label might be suppressed in order to display the table in one panel. Use the SAS system option LINESIZE= to specify a larger line size to accommodate variable labels. A shorter line size can break the table into two panels allowing labels to be displayed.
Displays the odds ratio estimates and the corresponding 95% Wald confidence intervals for variables that are not involved in nestings or interactions. For continuous explanatory variables, these odds ratios correspond to a unit increase in the risk factors. For more information, see the section Odds Ratio Estimation.
For more information, see the section Rank Correlation of Observed Responses and Predicted Probabilities.
Displays if you specify the CLPARM= option in the MODEL statement. For more information, see the section Confidence Intervals for Parameters.
Displays if you specify the ODDSRATIO statement for any effects with any class parameterizations. Also displays if you specify the CLODDS= option in the MODEL statement, except odds ratios are computed only for main effects not involved in interactions or nestings, and if the main effect is a CLASS variable, the parameterization must be EFFECT, REFERENCE, or GLM. For more information, see the section Odds Ratio Estimation.
Displays if you specify the COVB or CORRB option in the MODEL statement. For more information, see the section Iterative Algorithms for Model Fitting.
Displays the Wald test for each specified CONTRAST
statement. Small p-values reject ![]() . The "Coefficients of Contrast" table displays the contrast matrix if you specify the E
option, and the "Contrast Estimation and Testing Results by Row" table displays estimates and Wald tests for each row of
the contrast matrix if you specify the ESTIMATE=
option. For more information, see the sections CONTRAST Statement, Testing Linear Hypotheses about the Regression Coefficients, and Linear Predictor, Predicted Probability, and Confidence Limits.
. The "Coefficients of Contrast" table displays the contrast matrix if you specify the E
option, and the "Contrast Estimation and Testing Results by Row" table displays estimates and Wald tests for each row of
the contrast matrix if you specify the ESTIMATE=
option. For more information, see the sections CONTRAST Statement, Testing Linear Hypotheses about the Regression Coefficients, and Linear Predictor, Predicted Probability, and Confidence Limits.
Displays the Wald test for each specified TEST statement. For more information, see the sections Testing Linear Hypotheses about the Regression Coefficients and TEST Statement.
Displays if you specify the LACKFIT option in the MODEL statement. Small p-values reject the null hypothesis that the fitted model is adequate. The "Partition for the Hosmer and Lemeshow Test" table displays the grouping used in the test. For more information, see the section The Hosmer-Lemeshow Goodness-of-Fit Test.
Displays if you use the CTABLE option in the MODEL statement. If you specify a list of cutpoints with the PPROB= option , then the cutpoints are displayed in the Prob Level column. If you specify the prior event probabilities with the PEVENT= option, then the probabilities are displayed in the Prob Event column. The Correct column displays the number of correctly classified events and nonevents, the Incorrect Event column displays the number of nonevents incorrectly classified as events, and the Incorrect Nonevent column gives the number of events incorrectly classified as nonevents. For more information, see the section Classification Table.
Displays if you specify the INFLUENCE option in the MODEL statement. See the section Regression Diagnostics for more information about diagnostics from an unconditional analysis, and the section Regression Diagnostic Details for information about diagnostics from a conditional analysis.
Displays if you specify the FITSTAT option in the SCORE statement. For more information, see the section Scoring Data Sets.
Displayed if a ROC statement or a ROCCONTRAST statement is specified. For more information, about the Mann-Whitney statistics and the test and estimation computations, see the section ROC Computations. For more information about the other statistics, see the section Rank Correlation of Observed Responses and Predicted Probabilities.
The tables in this section are produced when the EXACT
statement is specified. If the METHOD=
NETWORKMC option is specified, the test and estimate tables are renamed "Monte Carlo" tables and a Monte Carlo standard error
column (![]() ) is displayed.
) is displayed.
-
Sufficient Statistics
Displays if you request an OUTDIST= data set in an EXACT statement. The table lists the parameters and their observed sufficient statistics. -
(Monte Carlo) Conditional Exact Tests
For more information, see the section Hypothesis Tests. -
(Monte Carlo) Exact Parameter Estimates
Displays if you specify the ESTIMATE option in the EXACT statement. This table gives individual parameter estimates for each variable (conditional on the values of all the other parameters in the model), confidence limits, and a two-sided p-value (twice the one-sided p-value) for testing that the parameter is zero. For more information, see the section Inference for a Single Parameter. -
(Monte Carlo) Exact Odds Ratios
Displays if you specify the ESTIMATE= ODDS or ESTIMATE= BOTH option in the EXACT statement. For more information, see the section Inference for a Single Parameter.