The HPNLMOD Procedure
MODEL dependent-variable ~ distribution ;
The MODEL statement is the mechanism for either using a distribution specification to specify the distribution of the data or using the RESIDUAL distribution to specify a predicted value. You must specify a single dependent variable from the input data set, a tilde (~), and then a distribution along with its parameters. You can specify the following values for distribution:
- RESIDUAL
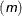 or LS
or LS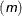
-
specifies no particular distribution. Instead the sum of squares of the differences between
 and the dependent variable is minimized.
and the dependent variable is minimized.
- NORMAL
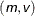
-
specifies a normal (Gaussian) distribution that has mean
 and variance
and variance  .
.
- BINARY
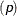
-
specifies a binary (Bernoulli) distribution that has probability
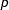 .
.
- BINOMIAL
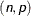
-
specifies a binomial distribution that has count
 and probability
and probability 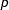 .
.
- GAMMA
- NEGBIN
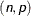
-
specifies a negative binomial distribution that has count
 and probability
and probability 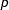 .
.
- POISSON
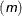
- GENERAL
-
specifies a general log-likelihood function that you construct by using SAS programming statements.
The MODEL statement must follow any SAS programming statements that you specify for computing parameters of the preceding distributions. For information about the built-in log-likelihood functions, see the section Built-In Log-Likelihood Functions .
