| The MDC Procedure |
Example 17.4 Testing for Homoscedasticity of the Utility Function
The conditional logit model imposes equal variances on random components of utility of all alternatives. This assumption can often be too restrictive and the calculated results misleading. This example shows several approaches to testing the homoscedasticity assumption.
The section Getting Started: MDC Procedure analyzes an HEV model by using Daganzo’s trinomial choice data and displays the HEV parameter estimates in Figure 17.15. The inverted scale estimates for mode "2" and mode "3" suggest that the conditional logit model (which imposes equal variances on random components of utility of all alternatives) might be misleading. The HEV estimation summary from that analysis is repeated in Output 17.4.1.
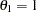 )
)
You can estimate the HEV model with unit scale restrictions on all three alternatives ( ) with the following statements. Output 17.4.2 displays the estimation summary.
) with the following statements. Output 17.4.2 displays the estimation summary.
/*-- HEV Estimation --*/
proc mdc data=newdata;
model decision = ttime /
type=hev
nchoice=3
hev=(unitscale=1 2 3, integrate=laguerre)
covest=hess;
id pid;
run;
 )
)
| Model Fit Summary | |
|---|---|
| Dependent Variable | decision |
| Number of Observations | 50 |
| Number of Cases | 150 |
| Log Likelihood | -34.12756 |
| Maximum Absolute Gradient | 6.7951E-9 |
| Number of Iterations | 5 |
| Optimization Method | Dual Quasi-Newton |
| AIC | 70.25512 |
| Schwarz Criterion | 72.16714 |
The test for scale equivalence (SCALE2=SCALE3=1) is performed using a likelihood ratio test statistic. The following SAS statements compute the test statistic (1.4276) and its  -value (0.4898) from the log-likelihood values in Figure 17.4.1 and Figure 17.4.2:
-value (0.4898) from the log-likelihood values in Figure 17.4.1 and Figure 17.4.2:
data _null_;
/*-- test for H0: scale2 = scale3 = 1 --*/
/* ln L(max) = -34.1276 */
/* ln L(0) = -33.4138 */
stat = -2 * ( - 34.1276 + 33.4138 );
df = 2;
p_value = 1 - probchi(stat, df);
put stat= p_value=;
run;
The test statistic fails to reject the null hypothesis of equal scale parameters, which implies that the random utility function is homoscedastic.
A multinomial probit model also allows heteroscedasticity of the random components of utility for different alternatives. Consider the following utility function:
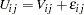 |
where
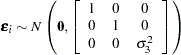 |
This multinomial probit model is estimated by using the following program. The estimation summary is displayed in Output 17.4.3.
/*-- Heteroscedastic Multinomial Probit --*/
proc mdc data=newdata;
model decision = ttime /
type=mprobit
nchoice=3
unitvariance=(1 2)
covest=hess;
id pid;
restrict RHO_31 = 0;
run;
| Model Fit Summary | |
|---|---|
| Dependent Variable | decision |
| Number of Observations | 50 |
| Number of Cases | 150 |
| Log Likelihood | -33.88604 |
| Log Likelihood Null (LogL(0)) | -54.93061 |
| Maximum Absolute Gradient | 5.60277E-6 |
| Number of Iterations | 8 |
| Optimization Method | Dual Quasi-Newton |
| AIC | 71.77209 |
| Schwarz Criterion | 75.59613 |
| Number of Simulations | 100 |
| Starting Point of Halton Sequence | 11 |
Next, the multinomial probit model with unit variances (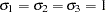 ) is estimated in the following statements. The estimation summary is displayed in Output 17.4.4.
) is estimated in the following statements. The estimation summary is displayed in Output 17.4.4.
/*-- Homoscedastic Multinomial Probit --*/
proc mdc data=newdata;
model decision = ttime /
type=mprobit
nchoice=3
unitvariance=(1 2 3)
covest=hess;
id pid;
restrict RHO_21 = 0;
run;
| Model Fit Summary | |
|---|---|
| Dependent Variable | decision |
| Number of Observations | 50 |
| Number of Cases | 150 |
| Log Likelihood | -34.54252 |
| Log Likelihood Null (LogL(0)) | -54.93061 |
| Maximum Absolute Gradient | 1.37303E-7 |
| Number of Iterations | 5 |
| Optimization Method | Dual Quasi-Newton |
| AIC | 71.08505 |
| Schwarz Criterion | 72.99707 |
| Number of Simulations | 100 |
| Starting Point of Halton Sequence | 11 |
The test for homoscedasticity ( = 1) under
= 1) under 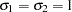 shows that the error variance is not heteroscedastic since the test statistic (
shows that the error variance is not heteroscedastic since the test statistic (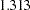 ) is less than
) is less than 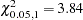 . The marginal probability or
. The marginal probability or  -value computed in the following program from the PROBCHI function is
-value computed in the following program from the PROBCHI function is 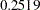 .
.
data _null_;
/*-- test for H0: sigma3 = 1 --*/
/* ln L(max) = -33.8860 */
/* ln L(0) = -34.5425 */
stat = -2 * ( -34.5425 + 33.8860 );
df = 1;
p_value = 1 - probchi(stat, df);
put stat= p_value=;
run;
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.