The HPCDM Procedure(Experimental)
Parameter Perturbation Analysis
It is important to realize that most of the parameters of the frequency and severity models are estimated and there is uncertainty associated with the parameter estimates. Any compound distribution estimate that is computed by using these uncertain parameter estimates is inherently uncertain. The aggregate loss sample that is simulated by using the mean estimates of the parameters is just one possible sample from the compound distribution. If information about parameter uncertainty is available, then it is recommended that you conduct parameter perturbation analysis that generates multiple samples of the compound distribution, in which each sample is simulated by using a set of perturbed parameter estimates. You can use the NPERTURBEDSAMPLES= option in the PROC HPCDM statement to specify the number of perturbed samples to be generated. The set of perturbed parameter estimates is created by making a random draw of the parameter values from their joint probability distribution. If you specify NPERTURBEDSAMPLES=P, then PROC HPCDM creates P sets of perturbed parameters and each set is used to simulate a full aggregate sample. The summary analysis of P such aggregate loss samples results in a set of P estimates for each summary statistic and percentile of the compound distribution. The mean and standard deviation of this set of P estimates quantify the uncertainty that is associated with the compound distribution.
The parameter uncertainty information is available in the form of either the variance-covariance matrix of the parameter estimates
or standard errors of the parameters estimates. If the variance-covariance matrix is available and is positive definite, then
PROC HPCDM assumes that the joint probability distribution of the parameter estimates is a multivariate normal distribution,
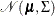 , where the mean vector
, where the mean vector 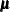 is the set of point parameter estimates and
is the set of point parameter estimates and 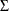 is the variance-covariance matrix. If the variance-covariance matrix is not available or is not positive definite, then PROC
HPCDM assumes that each parameter has a univariate normal distribution,
is the variance-covariance matrix. If the variance-covariance matrix is not available or is not positive definite, then PROC
HPCDM assumes that each parameter has a univariate normal distribution, 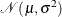 , where
, where 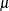 is the point estimate of the parameter and
is the point estimate of the parameter and  is the standard error of the parameter estimate.
is the standard error of the parameter estimate.
For severity models, the point parameter estimates are expected to be available in the SEVERITYEST= data set in observations for which _TYPE_='EST', the standard errors are expected to be available in the SEVERITYEST= data set in observations for which _TYPE_='STDERR', and the variance-covariance matrix is expected to be available in the SEVERITYEST= data set in observations for which _TYPE_='COV'. If you use the SEVERITY procedure to create the SEVERITYEST= data set, then you need to specify the COVOUT option in the PROC SEVERITY statement to make the variance-covariance estimates available in the SEVERITYEST= data set.
For the frequency model, you must use the COUNTREG procedure to create the COUNTSTORE= item store, which always contains the point estimates, standard errors, and variance-covariance matrix of the parameters.
If you specify the ADJUSTEDSEVERITY= option in the PROC HPCDM statement, then a separate perturbation analysis is conducted for the distribution of the aggregate adjusted loss.
