The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement
PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement -
Details
 Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsTweedie Distribution For Generalized Linear ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionResponse Level OrderingMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics
Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsTweedie Distribution For Generalized Linear ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionResponse Level OrderingMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics -
Examples
 Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson RegressionTweedie Regression
Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson RegressionTweedie Regression - References
Multinomial Models
This type of model applies to cases where an observation can fall into one of k categories. Binary data occur in the special case where k = 2. If there are 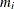 observations in a subpopulation i, then the probability distribution of the number falling into the k categories
observations in a subpopulation i, then the probability distribution of the number falling into the k categories 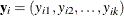 can be modeled by the multinomial distribution, defined in the section Response Probability Distributions, with
can be modeled by the multinomial distribution, defined in the section Response Probability Distributions, with 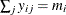 . The multinomial model is an ordinal model if the categories have a natural order.
. The multinomial model is an ordinal model if the categories have a natural order.
Residuals are not available in the OBSTATS table or the output data set for multinomial models.
By default, and consistently with binomial models, the GENMOD procedure orders the response categories for ordinal multinomial models from lowest to highest and models the probabilities of the lower response levels. You can change the way PROC GENMOD orders the response levels with the RORDER= option in the PROC GENMOD statement. The order that PROC GENMOD uses is shown in the "Response Profiles" output table described in the section Response Profile.
The GENMOD procedure supports only the ordinal multinomial model. If 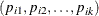 are the category probabilities, the cumulative category probabilities are modeled with the same link functions used for binomial
data. Let
are the category probabilities, the cumulative category probabilities are modeled with the same link functions used for binomial
data. Let 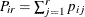 ,
, 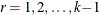 , be the cumulative category probabilities (note that
, be the cumulative category probabilities (note that 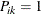 ). The ordinal model is
). The ordinal model is
![\[ g(P_{ir}) = \mu _ r + \mb{x}^\prime \bbeta ~ ~ ~ \mbox{for}~ ~ ~ r = 1, 2, \ldots , k\! -\! 1 \]](images/statug_genmod0313.png)
where 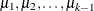 are intercept terms that depend only on the categories and
are intercept terms that depend only on the categories and 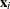 is a vector of covariates that does not include an intercept term. The logit, probit, and complementary log-log link functions
g are available. These are obtained by specifying the MODEL statement options DIST=MULTINOMIAL and LINK=CUMLOGIT (cumulative
logit), LINK=CUMPROBIT (cumulative probit), or LINK=CUMCLL (cumulative complementary log-log). Alternatively,
is a vector of covariates that does not include an intercept term. The logit, probit, and complementary log-log link functions
g are available. These are obtained by specifying the MODEL statement options DIST=MULTINOMIAL and LINK=CUMLOGIT (cumulative
logit), LINK=CUMPROBIT (cumulative probit), or LINK=CUMCLL (cumulative complementary log-log). Alternatively,
![\[ P_{ir} = \mr{F}(\mu _ r + \mb{x}^\prime \bbeta ) ~ ~ ~ \mbox{for}~ ~ ~ r = 1, 2, \ldots , k\! -\! 1 \]](images/statug_genmod0315.png)
where 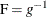 is a cumulative distribution function for the logistic, normal, or extreme-value distribution.
is a cumulative distribution function for the logistic, normal, or extreme-value distribution.
PROC GENMOD estimates the intercept parameters 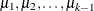 and regression parameters
and regression parameters 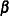 by maximum likelihood.
by maximum likelihood.
The subpopulations i are defined by constant values of the AGGREGATE= variable. This has no effect on the parameter estimates, but it does affect the deviance and Pearson chi-square statistics; it also affects parameter estimate standard errors if you specify the SCALE=DEVIANCE or SCALE=PEARSON option.