The GEE Procedure
Example 43.1 Comparison of the Marginal and Random Effect Models for Binary Data
A clinical trial (Stokes, Davis, and Koch 2012) was conducted to compare two treatments for a respiratory illness. Patients in each of two centers were randomly assigned to two groups: one group received the active treatment and one group received a placebo.
During treatment, respiratory status was determined for each of four visits and is represented by the variable Outcome (coded here as 0 = poor, 1 = good). The variables Center, Treatment, Sex, and Baseline (baseline respiratory status) are classification variables that have two levels. The variable Age (age at time of entry into the study) is a continuous variable.
All 111 patients completed the study. That is, there are no missing data for responses or covariates. The following statements
create the data set Resp:
data Resp; input Center ID Treatment $ Sex $ Age Baseline Visit1-Visit4; datalines; 1 1 P M 46 0 0 0 0 0 1 2 P M 28 0 0 0 0 0 1 3 A M 23 1 1 1 1 1 1 4 P M 44 1 1 1 1 0 1 5 P F 13 1 1 1 1 1 1 6 A M 34 0 0 0 0 0 ... more lines ... 2 51 A M 43 1 1 1 1 0 2 52 A F 39 0 1 1 1 1 2 53 A M 68 0 1 1 1 1 2 54 A F 63 1 1 1 1 1 2 55 A M 31 1 1 1 1 1 ;
data Resp; set Resp; Visit=1; Outcome=Visit1; output; Visit=2; Outcome=Visit2; output; Visit=3; Outcome=Visit3; output; Visit=4; Outcome=Visit4; output; run;
Suppose 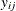 represents the respiratory status of patient i at the jth visit,
represents the respiratory status of patient i at the jth visit, 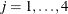 , and
, and 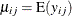 represents the mean of the respiratory status. Logistic regression is commonly used to analyze binary response data. You
can use the variance function for the binomial distribution,
represents the mean of the respiratory status. Logistic regression is commonly used to analyze binary response data. You
can use the variance function for the binomial distribution, 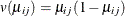 , and the logit link function,
, and the logit link function, 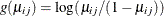 . The model for the mean is
. The model for the mean is 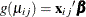 , where
, where 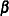 is a vector of regression parameters to be estimated.
is a vector of regression parameters to be estimated.
The following SAS statements perform the GEE model fit:
proc gee data=Resp descend;
class ID Treatment Center Sex Baseline;
model Outcome=Treatment Center Sex Age Baseline /
dist=bin link=logit;
repeated subject=ID(Center) / corr=exch corrw;
run;
Both the MODEL statement and the REPEATED statement are required.
In the MODEL statement, you use the DIST=BIN and LINK=LOGIT options to specify a logistic regression, and you specify Outcome as the response variable and Treatment, Center, Sex, Age, and Baseline as the explanatory variables. The DESCEND option in the PROC GEE statement requests that the probability that Outcome = 1 be modeled. If the DESCEND option had not been specified, the probability that Outcome = 0 would be modeled by default.
You use the REPEATED statement to specify the subject and the correlation structure of the responses. The SUBJECT=ID(CENTER)
option specifies that the observations in any single cluster are uniquely identified by Center and ID. An equivalent specification is SUBJECT=ID*CENTER. Because the same ID values are used in each center, one of these specifications is needed. If ID values were unique across all centers, SUBJECT=ID could be specified. The option TYPE=EXCH specifies the exchangeable working
correlation structure.
The "Model Information" table displayed in Output 43.1.1 provides information about the specified logistic regression model and the input data set.
Output 43.1.1: Model Information
General information about the GEE analysis is displayed in Output 43.1.2, and model fit criteria for the model are displayed in Output 43.1.3.
Output 43.1.2: Model Fitting Information
The results of GEE model fitting are displayed in Output 43.1.4. If you specify no other options, the standard errors, confidence intervals, Z scores, and p-values are based on empirical standard error estimates. You can specify the MODELSE option in the REPEATED statement to create a table that is based on model-based standard error estimates.
Output 43.1.4: Results of Model Fitting
| Parameter Estimates for Response Model | |||||||
|---|---|---|---|---|---|---|---|
| with Empirical Standard Error Estimates | |||||||
| Parameter | Estimate | Standard Error |
95% Confidence Limits | Z | Pr > |Z| | ||
| Intercept | 1.6391 | 0.5247 | 0.6107 | 2.6675 | 3.12 | 0.0018 | |
| Treatment | A | 1.2654 | 0.3467 | 0.5859 | 1.9448 | 3.65 | 0.0003 |
| Treatment | P | 0.0000 | 0.0000 | 0.0000 | 0.0000 | . | . |
| Center | 1 | -0.6495 | 0.3532 | -1.3418 | 0.0428 | -1.84 | 0.0660 |
| Center | 2 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | . | . |
| Sex | F | 0.1368 | 0.4402 | -0.7261 | 0.9996 | 0.31 | 0.7560 |
| Sex | M | 0.0000 | 0.0000 | 0.0000 | 0.0000 | . | . |
| Age | -0.0188 | 0.0130 | -0.0442 | 0.0067 | -1.45 | 0.1480 | |
| Baseline | 0 | -1.8457 | 0.3460 | -2.5238 | -1.1676 | -5.33 | <.0001 |
| Baseline | 1 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | . | . |
Treatment and Baseline appear to be strongly influential, and Center might be marginally significant.
For comparison, a generalized linear mixed model is fitted to the data set to obtain subject-specific effects. Specifically, consider the logistic regression model,
![\[ \mr{logit}(\mr{E}(y_{ij}|b_ i)) = {\mb{x}_{ij}}^\prime \bbeta ^* + b_ i \]](images/statug_gee0190.png)
where the random effect 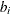 is normally distributed with zero mean and variance,
is normally distributed with zero mean and variance, 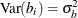 .
.
The following statements use the GLIMMIX procedure to fit a generalized linear mixed model:
proc glimmix data=Resp;
class ID Treatment Center Sex Baseline;
model Outcome (desc)=Treatment Center Sex Age Baseline /
dist=binary solution;
random ID(Center);
run;
Output 43.1.5 displays the parameter estimates for the fixed effects in the generalized linear mixed model.
Output 43.1.5: Parameter Estimates
| Solutions for Fixed Effects | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect | Treatment | Sex | Center | Baseline | Estimate | Standard Error |
DF | t Value | Pr > |t| |
| Intercept | 1.7936 | 0.6292 | 105 | 2.85 | 0.0053 | ||||
| Treatment | A | 1.4758 | 0.3898 | 333 | 3.79 | 0.0002 | |||
| Treatment | P | 0 | . | . | . | . | |||
| Center | 1 | -0.7201 | 0.4051 | 105 | -1.78 | 0.0784 | |||
| Center | 2 | 0 | . | . | . | . | |||
| Sex | F | 0.1732 | 0.5034 | 333 | 0.34 | 0.7310 | |||
| Sex | M | 0 | . | . | . | . | |||
| Age | -0.02011 | 0.01507 | 333 | -1.33 | 0.1831 | ||||
| Baseline | 0 | -2.1343 | 0.3971 | 333 | -5.38 | <.0001 | |||
| Baseline | 1 | 0 | . | . | . | . | |||
From Output 43.1.4 and Output 43.1.5, you can see that the parameter estimates from the marginal model and the mixed-effects model differ. For example, the estimated treatment effects are 1.2654 and 1.4758 from the marginal model and the mixed-effects model, respectively.
The interpretation of the model effects in the marginal and random models differs. For example, the estimated treatment effect
from the marginal model indicates that, on average, the odds of a good response for the patients is 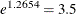 times higher when they receive the active treatment versus the placebo. The estimated treatment effect from the generalized
linear mixed model indicates that an individual patient’s odds of a good response is
times higher when they receive the active treatment versus the placebo. The estimated treatment effect from the generalized
linear mixed model indicates that an individual patient’s odds of a good response is 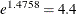 times higher when the patient receives the active treatment versus the placebo.
times higher when the patient receives the active treatment versus the placebo.
The choice of the marginal model or a subject-specific model often depends on the goal of your analysis: whether you are interested in population-averaged effects or subject-specific effects. For more information, see Diggle et al. (2002); Fitzmaurice, Laird, and Ware (2011).
