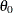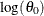The HPSEVERITY Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Predefined DistributionsCensoring and TruncationParameter Estimation MethodParameter InitializationEstimating Regression EffectsLevelization of Classification VariablesSpecification and Parameterization of Model EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDistributed and Multithreaded ComputationDefining a Severity Distribution Model with the FCMP ProcedurePredefined Utility FunctionsScoring FunctionsCustom Objective FunctionsInput Data SetsOutput Data SetsDisplayed OutputODS Graphics
Predefined DistributionsCensoring and TruncationParameter Estimation MethodParameter InitializationEstimating Regression EffectsLevelization of Classification VariablesSpecification and Parameterization of Model EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDistributed and Multithreaded ComputationDefining a Severity Distribution Model with the FCMP ProcedurePredefined Utility FunctionsScoring FunctionsCustom Objective FunctionsInput Data SetsOutput Data SetsDisplayed OutputODS Graphics -
Examples
 Defining a Model for Gaussian DistributionDefining a Model for the Gaussian Distribution with a Scale ParameterDefining a Model for Mixed-Tail DistributionsFitting a Scaled Tweedie Model with RegressorsFitting Distributions to Interval-Censored DataBenefits of Distributed and Multithreaded ComputingEstimating Parameters Using Cramér-von Mises EstimatorDefining a Finite Mixture Model That Has a Scale ParameterPredicting Mean and Value-at-Risk by Using Scoring FunctionsScale Regression with Rich Regression Effects
Defining a Model for Gaussian DistributionDefining a Model for the Gaussian Distribution with a Scale ParameterDefining a Model for Mixed-Tail DistributionsFitting a Scaled Tweedie Model with RegressorsFitting Distributions to Interval-Censored DataBenefits of Distributed and Multithreaded ComputingEstimating Parameters Using Cramér-von Mises EstimatorDefining a Finite Mixture Model That Has a Scale ParameterPredicting Mean and Value-at-Risk by Using Scoring FunctionsScale Regression with Rich Regression Effects - References
Scoring Functions
Scoring refers to the act of evaluating a distribution function, such as LOGPDF, SDF, or QUANTILE, on an observation by using the fitted parameter estimates of that distribution. You can do scoring in a DATA step by using the OUTEST= data set that you create with PROC HPSEVERITY. However, that approach requires some cumbersome programming. In order to simplify the scoring process, you can specify that PROC HPSEVERITY create scoring functions for each fitted distribution.
As an example, assume that you have fitted the Pareto distribution by using PROC HPSEVERITY and that it converges. Further assume that you want to use the fitted distribution to evaluate the probability of observing a loss value greater than some set of regulatory limits {L} that are encoded in a data set. You can simplify this scoring process as follows. First, in the PROC HPSEVERITY step that fits your distributions, you create the scoring functions library by specifying the OUTSCORELIB statement as illustrated in the following steps:
proc hpseverity data=input; loss lossclaim; dist pareto; outscorelib outlib=sasuser.fitdist; run;
Upon successful completion, if the Pareto distribution model has converged, then the Sasuser.Fitdist library contains the SEV_SDF scoring function in addition to other scoring functions, such as SEV_PDF, SEV_LOGPDF, and so on. Further, PROC HPSEVERITY also sets the CMPLIB system option to include the Sasuser.Fitdist library. If the set of limits {L} is recorded in the variable Limit in the scoring data set Work.Limits, then you can submit the following DATA step to compute the probability of seeing a loss greater than each limit:
data prob; set work.limits; exceedance_probability = sev_sdf(limit); run;
Without the use of scoring functions, you can still perform this scoring task, but the DATA step that you need to write to accomplish it becomes more complicated and less flexible. For example, you would need to read the parameter estimates from some output created by PROC HPSEVERITY. To do that, you would need to know the parameter names, which are different for different distributions; this in turn would require you to write a specific DATA step for each distribution or to write a SAS macro. With the use of scoring functions, you can accomplish that task much more easily.
If you fit multiple distributions, then you can specify the COMMONPACKAGE option in the OUTSCORELIB statement as follows:
proc hpseverity data=input; loss lossclaim; dist exp pareto weibull; outscorelib outlib=sasuser.fitdist commonpackage; run;
The preceding step creates scoring functions such as SEV_SDF_Exp, SEV_SDF_Pareto, and SEV_SDF_Weibull. You can use them to compare the probabilities of exceeding the limit for different distributions by using the following DATA step:
data prob; set work.limits; exceedance_exp = sev_sdf_exp(limit); exceedance_pareto = sev_sdf_pareto(limit); exceedance_weibull = sev_sdf_weibull(limit); run;
Formal Description
PROC HPSEVERITY creates a scoring function for each distribution function. A distribution function is defined as any function named dist_suffix, where dist is the name of a distribution that you specify in the DIST statement and the function’s signature is identical to the signature of the required distribution function such as dist_CDF or dist_LOGCDF. For example, for the function 'FOO_BAR' to be a distribution function, you must specify the distribution 'FOO' in the DIST statement and you must define 'FOO_BAR' in the following manner if the distribution 'FOO' has parameters named 'P1' and 'P2':
function FOO_BAR(y, P1, P2);
/* Code to compute BAR by using y, P1, and P2 */
R = <computed BAR>;
return (R);
endsub;
For more information about the signature that defines a distribution function, see the description of the dist_CDF function in the section Defining a Severity Distribution Model with the FCMP Procedure.
The name and package of the scoring function of a distribution function depend on whether you specify the COMMONPACKAGE option in the OUTSCORELIB statement.
When you do not specify the COMMONPACKAGE option, the scoring function that corresponds to the distribution function dist_suffix is named SEV_suffix, where SEV_ is the standard prefix of all scoring functions. The scoring function is created in a package named dist. Each scoring function accepts only one argument, the value of the loss variable, and returns the same value as the value returned by the corresponding distribution function for the final estimates of the distribution’s parameters. For example, for the preceding 'FOO_BAR' distribution function, the scoring function named 'SEV_BAR' is created in the package named 'FOO' and 'SEV_BAR' has the following signature:
function SEV_BAR(y);
/* returns value of FOO_BAR for the supplied value
of y and fitted values of P1, P2 */
endsub;
If you specify the COMMONPACKAGE option in the OUTSCORELIB statement, then the scoring function that corresponds to the distribution function dist_suffix is named SEV_suffix_dist, where SEV_ is the standard prefix of all scoring functions. The scoring function is created in a package named sevfit. For example, for the preceding 'FOO_BAR' distribution function, if you specify the COMMONPACKAGE option, the scoring function named 'SEV_BAR_FOO' is created in the sevfit package and 'SEV_BAR_FOO' has the following signature:
function SEV_BAR_FOO(y);
/* returns value of FOO_BAR for the supplied value
of y and fitted values of P1, P2 */
endsub;
Scoring Functions for the Scale Regression Model
If you use the SCALEMODEL statement to specify a scale regression model, then PROC HPSEVERITY generates the scoring functions when you specify only singleton continuous effects. If you specify interaction or classification effects, then scoring functions are not generated.
For a scale regression model, the estimate of the scale parameter or the log-transformed scale parameter of the distribution depends on the values of the regressors. So PROC HPSEVERITY creates a scoring function that has the following signature, where x{*} represents the array of regressors:
function SEV_BAR(y, x{*});
/* returns value of FOO_BAR for the supplied value of x and fitted values of P1, P2 */
endsub;
As an illustration of using this form, assume that you submit the following PROC HPSEVERITY step to create the scoring library
Sasuser.Scalescore:
proc hpseverity data=input; loss lossclaim; scalemodel x1-x3; dist pareto; outscorelib outlib=sasuser.scalescore; run;
Your scoring data set must contain all the regressors that you specify in the SCALEMODEL statement. You can submit the following DATA step to score observations by using the scale regression model:
data prob;
array regvals{*} x1-x3;
set work.limits;
exceedance_probability = sev_sdf(limit, regvals);
run;
PROC HPSEVERITY creates two utility functions, SEV_NUMREG and SEV_REGNAME, in the OUTLIB= library that return the number of regressors and name of a given regressor, respectively. They are described in detail in the next section. These utility functions are useful when you do not have easy access to the regressor names in the SCALEMODEL statement. You can use the utility functions as follows:
data prob;
array regvals{10} _temporary_;
set work.limits;
do i = 1 to sev_numreg();
regvals(i) = input(vvaluex(sev_regname(i)), best12.);
end;
exceedance_probability = sev_sdf(limit, regvals);
run;
The dimension of the regressor values array that you supply to the scoring function must be equal to 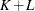 , where K is the number of regressors that you specify in the SCALEMODEL statement irrespective of whether PROC HPSEVERITY deems any
of those regressors to be redundant. L is 1 if you specify an OFFSET=
variable in the SCALEMODEL statement, and 0 otherwise.
, where K is the number of regressors that you specify in the SCALEMODEL statement irrespective of whether PROC HPSEVERITY deems any
of those regressors to be redundant. L is 1 if you specify an OFFSET=
variable in the SCALEMODEL statement, and 0 otherwise.
Utility Functions and Subroutines in the OUTLIB= Library
In addition to creating the scoring functions for all distribution functions, PROC HPSEVERITY creates the following utility functions and subroutines in the OUTLIB= library.
If you use the SCALEMODEL statement to specify a scale regression model, and if you specify only singleton continuous effects, then the following helper functions and subroutines are also created in the OUTLIB= library.