-
ALGORITHM=algorithm <( suboptions )>
-
specifies an algorithm for estimating the regression parameters. Three algorithms are available: simplex (SIMPLEX), interior point (INTERIOR), and smoothing (SMOOTH).
The default algorithm depends on the number of observations (n) and the number of covariates (p) in the model estimation. See Table 83.2 for the relevant defaults.
Table 83.2: The Default Estimation Algorithm
| |
|
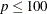
|
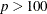
|
|
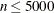
|
|
Simplex
|
Smoothing
|
|
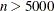
|
|
Interior point
|
Smoothing
|
Table 83.3 summarizes the options available for each of these methods.
Table 83.3: Options for Estimation Algorithms
|
algorithm
|
|
Algorithm
|
suboption
|
|
INTERIOR
|
|
Interior point
|
KAPPA=, MAXIT=, and TOLERANCE=
|
|
SIMPLEX
|
|
Simplex
|
MAXSTATIONARY=
|
|
SMOOTH
|
|
Smoothing
|
RRATIO=
|
If you specify ALGORITHM=SIMPLEX, you can specify the following suboption:
- MAXSTATIONARY=m
-
requests that the algorithm terminate if the objective function has not improved for m consecutive iterations. By default, m = 1000.
If you specify ALGORITHM=INTERIOR, you can specify the following suboptions:
- KAPPA=value
-
specifies the step-length parameter for the interior point algorithm. This parameter should be between 0 and 1. The larger
the parameter, the faster the algorithm. However, numeric instability can occur as the parameter approaches 1. By default,
KAPPA=0.99995. For more information, see the section Interior Point Algorithm.
- MAXIT=m
-
sets the maximum number of iterations for the interior point algorithm. By default, MAXIT=1000.
- TOLERANCE=value
-
specifies the tolerance for the convergence criterion of the interior point algorithm. By default, TOLERANCE=1E–8. The QUANTREG
procedure uses the duality gap as the convergence criterion. For more information, see the section Interior Point Algorithm.
If you specify ALGORITHM=INTERIOR, you can also use the PERFORMANCE statement to enable parallel computing when multiple processors
are available in the hardware.
If you specify ALGORITHM=SMOOTH, you can specify the following suboption:
- RRATIO=value
-
specifies the reduction ratio for the smoothing algorithm. This ratio is used to reduce the threshold of the smoothing algorithm.
The value should be between 0 and 1. In theory, the smaller the reduction ratio, the faster the smoothing algorithm. However, in practice,
the optimal ratio is quite dependent on the data. For more information, see the section Smoothing Algorithm.
-
ALPHA=value
-
specifies the level of significance  for
for 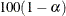 % confidence intervals for regression parameters. The value must be between 0 and 1. The default is ALPHA=0.05, which corresponds to a 0.95 confidence interval.
% confidence intervals for regression parameters. The value must be between 0 and 1. The default is ALPHA=0.05, which corresponds to a 0.95 confidence interval.
-
CI=NONE | RANK | SPARSITY<(BF | HS)></IID> | RESAMPLING<(NREP=n)>
-
specifies a method for computing confidence intervals for regression parameters. When you specify CI=SPARSITY or CI=RESAMPLING, the QUANTREG procedure also computes standard errors, t values, and p-values for regression parameters.
The following table summarizes these methods.
Table 83.4: Options for Confidence Intervals
|
Value of CI=
|
|
Method
|
Additional Options
|
|
NONE
|
|
No confidence intervals computed
|
|
|
RANK
|
|
By inverting rank-score tests
|
|
|
RESAMPLING
|
|
By resampling
|
NREP
|
|
SPARSITY
|
|
By estimating sparsity function
|
HS, BF, and IID
|
By default, when there are fewer than 5,000 observations, fewer than 20 variables in the data set, and the algorithm is simplex,
the QUANTREG procedure computes confidence intervals by using the inverted rank-score test method. Otherwise, the resampling
method is used.
By default, confidence intervals are not computed for the quantile process, which is estimated when you specify the QUANTILE=PROCESS
option in the MODEL statement. Confidence intervals for the quantile process are computed by using the sparsity or resampling
methods when you specify CI=SPARSITY or CI=RESAMPLING, respectively. The rank method for confidence intervals is not available
for quantile processes because it is computationally prohibitive.
When you specify the SPARSITY option, you have two suboptions for estimating the sparsity function. If you specify the IID
suboption, the sparsity function is estimated by assuming that the errors in the linear model are independent and identically
distributed (iid). By default, the sparsity function is estimated by assuming that the conditional quantile function is locally
linear. For more information, see the section Sparsity. For both methods, two bandwidth selection methods are available: You can specify the BF suboption for the Bofinger method
or the the HS suboption for the Hall-Sheather method. By default, the Hall-Sheather method is used.
When you specify the RESAMPLING option, you can specify the NREP=n suboption for the number of repetitions. By default, NREP=200. The value of n must be greater than 50.
-
DATA=SAS-data-set
-
specifies the input SAS data set to be used by the QUANTREG procedure. By default, the most recently created SAS data set is used.
-
INEST=SAS-data-set
-
specifies an input SAS data set that contains initial estimates for all the parameters in the model. The interior point algorithm and the smoothing algorithm use these estimates as a start. For a detailed
description of the contents of the INEST= data set, see the section INEST= Data Set.
-
NAMELEN=n
-
restricts the length of effect names in tables and output data sets to n characters, where n is a value between 20 and 200. By default, NAMELEN=20.
-
ORDER=DATA | FORMATTED | FREQ | INTERNAL
-
specifies the sort order for the levels of the classification variables (which are specified in the CLASS
statement).
This option applies to the levels for all classification variables, except when you use the (default) ORDER=FORMATTED option
with numeric classification variables that have no explicit format. In that case, the levels of such variables are ordered
by their internal value.
The ORDER= option can take the following values:
|
Value of ORDER=
|
Levels Sorted By
|
|
DATA
|
Order of appearance in the input data set
|
|
FORMATTED
|
External formatted value, except for numeric variables with no explicit format, which are sorted by their unformatted (internal)
value
|
|
FREQ
|
Descending frequency count; levels with the most observations come first in the order
|
|
INTERNAL
|
Unformatted value
|
By default, ORDER=FORMATTED. For ORDER=FORMATTED and ORDER=INTERNAL, the sort order is machine-dependent.
For more information about sort order, see the chapter on the SORT procedure in the
Base SAS Procedures Guide and the discussion of BY-group processing in
SAS Language Reference: Concepts.
-
OUTEST=SAS-data-set
-
specifies an output SAS data set to contain the parameter estimates for all quantiles. See the section OUTEST= Data Set for a detailed description of the contents of the OUTEST= data set.
-
PLOT | PLOTS<(global-plot-options)> <=plot-request>
PLOT | PLOTS<(global-plot-options)> <=(plot-request < …plot-request > )>
-
specifies options that control details of the plots. These plots fall into two categories: diagnostic plots and fit plots. You can also use the PLOT=
option in the MODEL statement to request the quantile process plot for any effects that are specified in the model. If you
do not specify the PLOTS= option, PROC QUANTREG produces the quantile fit plot by default when a single continuous variable
is specified in the model.
When you specify only one plot-request, you can omit the parentheses around the plot request.
Here are some examples:
plots=ddplot
plots=(ddplot rdplot)
ODS Graphics must be enabled before plots can be requested. For example:
ods graphics on;
proc quantreg plots=fitplot;
model y=x1;
run;
ods graphics off;
For more information about enabling and disabling ODS Graphics, see the section Enabling and Disabling ODS Graphics in Chapter 21: Statistical Graphics Using ODS.
You can specify the following global-plot-options, which apply to all plots that PROC QUANTREG generates:
-
MAXPOINTS=NONE | number
-
suppresses plots that have elements that require processing more than number points. The default is MAXPOINTS=5000. This cutoff is ignored if you specify MAXPOINTS=NONE.
-
ONLY
-
suppresses the default quantile fit plot. Only plots specifically requested are displayed.
You can specify the following plot-requests:
-
ALL
-
creates all appropriate plots.
-
DDPLOT<(LABEL=ALL | LEVERAGE | NONE | OUTLIER)>
-
creates a plot of robust distance against Mahalanobis distance. For more information about robust distance, see the section
Leverage Point and Outlier Detection. The LABEL= option specifies how the points on this plot are to be labeled, as summarized by Table 83.5.
Table 83.5: Options for Label
|
Value of LABEL=
|
|
Label Method
|
|
ALL
|
|
Label all points
|
|
LEVERAGE
|
|
Label leverage points
|
|
NONE
|
|
No labels
|
|
OUTLIERS
|
|
Label outliers
|
By default, the QUANTREG procedure labels both outliers and leverage points.
If you specify ID variables in the ID statement, the values of the first ID variable are used as labels; otherwise, observation
numbers are used as labels.
-
FITPLOT<(NOLIMITS | SHOWLIMITS | NODATA)>
-
creates a plot of fitted conditional quantiles against the single continuous variable that is specified in the model. This
plot is produced only when the response is modeled as a function of a single continuous variable. Multiple lines or curves
are drawn on this plot if you specify several quantiles with the QUANTILE= option in the MODEL statement. By default, confidence
limits are added to the plot when a single quantile is requested, and the confidence limits are not shown on the plot when
multiple quantiles are requested. The NOLIMITS option suppresses the display of the confidence limits. The SHOWLIMITS option
adds the confidence limits when multiple quantiles are requested. The NODATA option suppresses the display of the observed
data, which are superimposed on the plot by default.
-
HISTOGRAM
-
creates a histogram (based on the quantile regression estimates) for the standardized residuals. The histogram is superimposed
with a normal density curve and a kernel density curve.
-
NONE
-
suppresses all plots.
-
QQPLOT
-
creates the normal quantile-quantile plot (based on the quantile regression estimates) for the standardized residuals.
-
RDPLOT<(LABEL=ALL | LEVERAGE | NONE | OUTLIER)>
-
creates the plot of standardized residual against robust distance. For more information about robust distance, see the section
Leverage Point and Outlier Detection.
The LABEL= option specifies a label method for points on this plot. These label methods are described in Table 83.5.
By default, the QUANTREG procedure labels both outliers and leverage points.
If you specify ID variables in the ID statement, the values of the first ID variable are used as labels; otherwise, observation
numbers are used as labels.
-
PP
-
requests preprocessing to speed up the interior point algorithm or the smoothing algorithm. The preprocessing uses a subsampling algorithm (which assumes that the data set is evenly distributed)
to iteratively reduce the original problem to a smaller one. Preprocessing should be used only for very large data sets, such
as data sets with more than 100,000 observations. For more information, see Portnoy and Koenker (1997).
