The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement
PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement -
Details
 Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics
Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics -
Examples
 Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson Regression
Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson Regression - References
Let ![]() ,
, ![]() ,
, ![]() , represent the jth measurement on the ith subject. There are
, represent the jth measurement on the ith subject. There are ![]() measurements on subject i and
measurements on subject i and ![]() total measurements.
total measurements.
Correlated data are modeled using the same link function and linear predictor setup (systematic component) as the independence
case. The random component is described by the same variance functions as in the independence case, but the covariance structure
of the correlated measurements must also be modeled. Let the vector of measurements on the ith subject be ![]() with corresponding vector of means
with corresponding vector of means ![]() , and let
, and let ![]() be the covariance matrix of
be the covariance matrix of ![]() . Let the vector of independent, or explanatory, variables for the jth measurement on the ith subject be
. Let the vector of independent, or explanatory, variables for the jth measurement on the ith subject be
The generalized estimating equation of Liang and Zeger (1986) for estimating the ![]() vector of regression parameters
vector of regression parameters ![]() is an extension of the independence estimating equation to correlated data and is given by
is an extension of the independence estimating equation to correlated data and is given by
where
Since
where g is the link function, the ![]() matrix of partial derivatives of the mean with respect to the regression parameters for the ith subject is given by
matrix of partial derivatives of the mean with respect to the regression parameters for the ith subject is given by
![\[ \mb {D}^\prime _ i = \frac{\partial \bmu _{i}^{\prime }}{\partial \bbeta } = \left[\begin{array}{ccc} \displaystyle \frac{x_{i11}}{g^{\prime }(\mu _{i1})} & \ldots & \displaystyle \frac{x_{in_ i1}}{g^{\prime }(\mu _{in_ i})} \\ \vdots & & \vdots \\ \displaystyle \frac{x_{i1p}}{g^{\prime }(\mu _{i1})} & \ldots & \displaystyle \frac{x_{in_ ip}}{g^{\prime }(\mu _{in_ i})} \\ \end{array} \right] \]](images/statug_genmod0395.png)
Let ![]() be an
be an ![]() “working” correlation matrix that is fully specified by the vector of parameters
“working” correlation matrix that is fully specified by the vector of parameters ![]() . The covariance matrix of
. The covariance matrix of ![]() is modeled as
is modeled as
where ![]() is an
is an ![]() diagonal matrix with
diagonal matrix with ![]() as the jth diagonal element and
as the jth diagonal element and ![]() is an
is an ![]() diagonal matrix with
diagonal matrix with ![]() as the jth diagonal, where
as the jth diagonal, where ![]() is a weight specified with the WEIGHT statement. If there is no WEIGHT statement,
is a weight specified with the WEIGHT statement. If there is no WEIGHT statement, ![]() for all i and j. If
for all i and j. If ![]() is the true correlation matrix of
is the true correlation matrix of ![]() , then
, then ![]() is the true covariance matrix of
is the true covariance matrix of ![]() .
.
The working correlation matrix is usually unknown and must be estimated. It is estimated in the iterative fitting process
by using the current value of the parameter vector ![]() to compute appropriate functions of the Pearson residual
to compute appropriate functions of the Pearson residual
If you specify the working correlation as ![]() , which is the identity matrix, the GEE reduces to the independence estimating equation.
, which is the identity matrix, the GEE reduces to the independence estimating equation.
Following are the structures of the working correlation supported by the GENMOD procedure and the estimators used to estimate the working correlations.
|
Working Correlation Structure |
Estimator |
|
|---|---|---|
|
Fixed |
||
|
|
The working correlation is not estimated in this case. |
|
|
Independent |
||
|
|
The working correlation is not estimated in this case. |
|
|
|
||
|
|
|
|
|
|
||
|
Exchangeable |
||
|
|
|
|
|
|
||
|
Unstructured |
||
|
|
|
|
|
Autoregressive AR(1) |
||
|
|
|
|
|
|
||
The dispersion parameter ![]() is estimated by
is estimated by
where ![]() is the total number of measurements and p is the number of regression parameters.
is the total number of measurements and p is the number of regression parameters.
The square root of ![]() is reported by PROC GENMOD as the scale parameter in the “Analysis of GEE Parameter Estimates Model-Based Standard Error Estimates” output table. If a fixed scale parameter is specified with the NOSCALE option in the MODEL statement, then the fixed value
is used in estimating the model-based covariance matrix and standard errors.
is reported by PROC GENMOD as the scale parameter in the “Analysis of GEE Parameter Estimates Model-Based Standard Error Estimates” output table. If a fixed scale parameter is specified with the NOSCALE option in the MODEL statement, then the fixed value
is used in estimating the model-based covariance matrix and standard errors.
The following is an algorithm for fitting the specified model by using GEEs. Note that this is not in general a likelihood-based method of estimation, so that inferences based on likelihoods are not possible for GEE methods.
-
Compute an initial estimate of
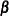 with an ordinary generalized linear model assuming independence.
with an ordinary generalized linear model assuming independence.
-
Compute the working correlations
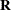 based on the standardized residuals, the current
based on the standardized residuals, the current 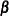 , and the assumed structure of
, and the assumed structure of 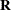 .
.
-
Compute an estimate of the covariance:
![\[ \mb {V}_{i} = \phi \mb {A}_ i^{\frac{1}{2}} \mb {W}_ i^{-\frac{1}{2}} \hat{\mb {R}}(\balpha ) \mb {W}_ i^{-\frac{1}{2}} \mb {A}_ i^{\frac{1}{2}} \]](images/statug_genmod0427.png)
-
Update
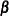 :
:
![\[ \bbeta _{r+1} = \bbeta _{r} + \left[\sum _{i=1}^{K} \frac{\partial \bmu _ i}{\partial \bbeta }^{\prime } \mb {V}_{i}^{-1} \frac{\partial \bmu _ i}{\partial \bbeta } \right]^{-1} \left[ \sum _{i=1}^ K \frac{\partial \bmu _ i}{\partial \bbeta }^{\prime }\mb {V}_{i}^{-1} (\mb {Y}_{i}-\bmu _ i) \right] \]](images/statug_genmod0428.png)
-
Repeat steps 2-4 until convergence.
See Diggle, Liang, and Zeger (1994, Chapter 11) for a discussion of missing values in longitudinal data. Suppose that you intend to take measurements ![]() for the ith unit. Missing values for which
for the ith unit. Missing values for which ![]() are missing whenever
are missing whenever ![]() is missing for all
is missing for all ![]() are called dropouts. Otherwise, missing values that occur intermixed with nonmissing values are intermittent missing values. The GENMOD procedure can estimate the working correlation from data containing both types of missing values
by using the all available pairs method, in which all nonmissing pairs of data are used in the moment estimators of the working correlation parameters defined
previously. The resulting covariances and standard errors are valid under the missing completely at random (MCAR) assumption.
are called dropouts. Otherwise, missing values that occur intermixed with nonmissing values are intermittent missing values. The GENMOD procedure can estimate the working correlation from data containing both types of missing values
by using the all available pairs method, in which all nonmissing pairs of data are used in the moment estimators of the working correlation parameters defined
previously. The resulting covariances and standard errors are valid under the missing completely at random (MCAR) assumption.
For example, for the unstructured working correlation model,
where the sum is over the units that have nonmissing measurements at times j and k, and ![]() is the number of units with nonmissing measurements at j and k. Estimates of the parameters for other working correlation types are computed in a similar manner, using available nonmissing
pairs in the appropriate moment estimators.
is the number of units with nonmissing measurements at j and k. Estimates of the parameters for other working correlation types are computed in a similar manner, using available nonmissing
pairs in the appropriate moment estimators.
The contribution of the ith unit to the parameter update equation is computed by omitting the elements of ![]() , the columns of
, the columns of ![]() , and the rows and columns of
, and the rows and columns of ![]() corresponding to missing measurements.
corresponding to missing measurements.
The model-based estimator of ![]() is given by
is given by
where
This is the GEE equivalent of the inverse of the Fisher information matrix that is often used in generalized linear models
as an estimator of the covariance estimate of the maximum likelihood estimator of ![]() . It is a consistent estimator of the covariance matrix of
. It is a consistent estimator of the covariance matrix of ![]() if the mean model and the working correlation matrix are correctly specified.
if the mean model and the working correlation matrix are correctly specified.
The estimator
is called the empirical, or robust, estimator of the covariance matrix of ![]() , where
, where
It has the property of being a consistent estimator of the covariance matrix of ![]() , even if the working correlation matrix is misspecified—that is, if
, even if the working correlation matrix is misspecified—that is, if ![]() . For further information about the robust variance estimate, see Zeger, Liang, and Albert (1988); Royall (1986); White (1982). In computing
. For further information about the robust variance estimate, see Zeger, Liang, and Albert (1988); Royall (1986); White (1982). In computing ![]() ,
, ![]() and
and ![]() are replaced by estimates, and
are replaced by estimates, and ![]() is replaced by the estimate
is replaced by the estimate
Lipsitz, Kim, and Zhao (1994) and Miller, Davis, and Landis (1993) describe how to extend GEEs to multinomial data. Currently, only the independent working correlation is available for multinomial models in PROC GENMOD.
If the responses are binary (that is, they take only two values), then there is an alternative method to account for the association among the measurements. The alternating logistic regressions (ALR) algorithm of Carey, Zeger, and Diggle (1993) models the association between pairs of responses with log odds ratios, instead of with correlations, as ordinary GEEs do.
For binary data, the correlation between the jth and kth response is, by definition,
The joint probability in the numerator satisfies the following bounds, by elementary properties of probability, since ![]() :
:
The correlation, therefore, is constrained to be within limits that depend in a complicated way on the means of the data.
The odds ratio, defined as
is not constrained by the means and is preferred, in some cases, to correlations for binary data.
The ALR algorithm seeks to model the logarithm of the odds ratio, ![]() , as
, as
where ![]() is a
is a ![]() vector of regression parameters and
vector of regression parameters and ![]() is a fixed, specified vector of coefficients.
is a fixed, specified vector of coefficients.
The parameter ![]() can take any value in
can take any value in ![]() with
with ![]() corresponding to no association.
corresponding to no association.
The log odds ratio, when modeled in this way with a regression model, can take different values in subgroups defined by ![]() . For example,
. For example, ![]() can define subgroups within clusters, or it can define “block effects” between clusters.
can define subgroups within clusters, or it can define “block effects” between clusters.
You specify a GEE model for binary data that uses log odds ratios by specifying a model for the mean, as in ordinary GEEs,
and a model for the log odds ratios. You can use any of the link functions appropriate for binary data in the model for the
mean, such as logistic, probit, or complementary log-log. The ALR algorithm alternates between a GEE step to update the model
for the mean and a logistic regression step to update the log odds ratio model. Upon convergence, the ALR algorithm provides
estimates of the regression parameters for the mean, ![]() , the regression parameters for the log odds ratios,
, the regression parameters for the log odds ratios, ![]() , their standard errors, and their covariances.
, their standard errors, and their covariances.
Specifying a regression model for the log odds ratio requires you to specify rows of the ![]() matrix
matrix ![]() for each cluster i and each unique within-cluster pair
for each cluster i and each unique within-cluster pair ![]() . The GENMOD procedure provides several methods of specifying
. The GENMOD procedure provides several methods of specifying ![]() . These are controlled by the LOGOR=keyword and associated options in the REPEATED statement. The supported keywords and the resulting log odds ratio models are described
as follows.
. These are controlled by the LOGOR=keyword and associated options in the REPEATED statement. The supported keywords and the resulting log odds ratio models are described
as follows.
- EXCH
-
specifies exchangeable log odds ratios. In this model, the log odds ratio is a constant for all clusters i and pairs
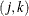 . The parameter
. The parameter  is the common log odds ratio.
is the common log odds ratio.
![\[ \mb {z}_{ijk} = 1 \; \; \mbox{ for all } \; \; i, j, k \]](images/statug_genmod0459.png)
- FULLCLUST
-
specifies fully parameterized clusters. Each cluster is parameterized in the same way, and there is a parameter for each unique pair within clusters. If a complete cluster is of size n, then there are
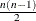 parameters in the vector
parameters in the vector  . For example, if a full cluster is of size 4, then there are
. For example, if a full cluster is of size 4, then there are 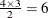 parameters, and the
parameters, and the  matrix is of the form
matrix is of the form
![\[ \bZ = \left[ \begin{array}{cccccc} 1 & 0 & 0 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 & 0 & 0 \\ 0 & 0 & 1 & 0 & 0 & 0 \\ 0 & 0 & 0 & 1 & 0 & 0 \\ 0 & 0 & 0 & 0 & 1 & 0 \\ 0 & 0 & 0 & 0 & 0 & 1 \\ \end{array} \right] \]](images/statug_genmod0462.png)
The elements of
 correspond to log odds ratios for cluster pairs in the following order:
correspond to log odds ratios for cluster pairs in the following order:
Pair
Parameter
(1,2)
Alpha1
(1,3)
Alpha2
(1,4)
Alpha3
(2.3)
Alpha4
(2,4)
Alpha5
(3,4)
Alpha6
- LOGORVAR(variable)
-
specifies log odds ratios by cluster. The argument variable is a variable name that defines the “block effects” between clusters. The log odds ratios are constant within clusters, but they take a different value for each different value of the variable. For example, if
Centeris a variable in the input data set taking a different value for k treatment centers, then specifying LOGOR=LOGORVAR(Center) requests a model with different log odds ratios for each of the k centers, constant within center. - NESTK
-
specifies k-nested log odds ratios. You must also specify the SUBCLUST=variable option to define subclusters within clusters. Within each cluster, PROC GENMOD computes a log odds ratio parameter for pairs having the same value of variable for both members of the pair and one log odds ratio parameter for each unique combination of different values of variable.
- NEST1
-
specifies 1-nested log odds ratios. You must also specify the SUBCLUST=variable option to define subclusters within clusters. There are two log odds ratio parameters for this model. Pairs having the same value of variable correspond to one parameter; pairs having different values of variable correspond to the other parameter. For example, if clusters are hospitals and subclusters are wards within hospitals, then patients within the same ward have one log odds ratio parameter, and patients from different wards have the other parameter.
- ZFULL
-
specifies the full
 matrix. You must also specify a SAS data set containing the
matrix. You must also specify a SAS data set containing the  matrix with the ZDATA=data-set-name option. Each observation in the data set corresponds to one row of the
matrix with the ZDATA=data-set-name option. Each observation in the data set corresponds to one row of the  matrix. You must specify the ZDATA data set as if all clusters are complete—that is, as if all clusters are the same size
and there are no missing observations. The ZDATA data set has
matrix. You must specify the ZDATA data set as if all clusters are complete—that is, as if all clusters are the same size
and there are no missing observations. The ZDATA data set has ![$K [n_{\mathit{max}}(n_{\mathit{max}}-1)/2]$](images/statug_genmod0463.png) observations, where K is the number of clusters and
observations, where K is the number of clusters and 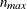 is the maximum cluster size. If the members of cluster i are ordered as
is the maximum cluster size. If the members of cluster i are ordered as 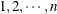 , then the rows of the
, then the rows of the  matrix must be specified for pairs in the order
matrix must be specified for pairs in the order 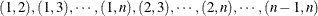 . The variables specified in the REPEATED statement for the SUBJECT effect must also be present in the ZDATA= data set to
identify clusters. You must specify variables in the data set that define the columns of the
. The variables specified in the REPEATED statement for the SUBJECT effect must also be present in the ZDATA= data set to
identify clusters. You must specify variables in the data set that define the columns of the  matrix by the ZROW=variable-list option. If there are q columns (q variables in variable-list), then there are q log odds ratio parameters. You can optionally specify variables indicating the cluster pairs corresponding to each row of
the
matrix by the ZROW=variable-list option. If there are q columns (q variables in variable-list), then there are q log odds ratio parameters. You can optionally specify variables indicating the cluster pairs corresponding to each row of
the  matrix with the YPAIR=(variable1, variable2) option. If you specify this option, the data from the ZDATA data set are sorted within each cluster by variable1 and variable2. See Example 40.6 for an example of specifying a full
matrix with the YPAIR=(variable1, variable2) option. If you specify this option, the data from the ZDATA data set are sorted within each cluster by variable1 and variable2. See Example 40.6 for an example of specifying a full  matrix.
matrix.
- ZREP
-
specifies a replicated
 matrix. You specify
matrix. You specify  matrix data exactly as you do for the ZFULL case, except that you specify only one complete cluster. The
matrix data exactly as you do for the ZFULL case, except that you specify only one complete cluster. The  matrix for the one cluster is replicated for each cluster. The number of observations in the ZDATA data set is
matrix for the one cluster is replicated for each cluster. The number of observations in the ZDATA data set is 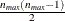 , where
, where 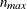 is the size of a complete cluster (a cluster with no missing observations).
is the size of a complete cluster (a cluster with no missing observations).
- ZREP(matrix)
-
specifies direct input of the replicated
 matrix. You specify the
matrix. You specify the  matrix for one cluster with the syntax LOGOR=ZREP (
matrix for one cluster with the syntax LOGOR=ZREP ( 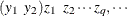 ), where
), where 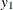 and
and 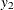 are numbers representing a pair of observations and the values
are numbers representing a pair of observations and the values 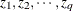 make up the corresponding row of the
make up the corresponding row of the  matrix. The number of rows specified is
matrix. The number of rows specified is 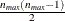 , where
, where 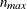 is the size of a complete cluster (a cluster with no missing observations). For example,
is the size of a complete cluster (a cluster with no missing observations). For example,
logor = zrep((1 2) 1 0, (1 3) 1 0, (1 4) 1 0, (2 3) 1 1, (2 4) 1 1, (3 4) 1 1)specifies the
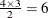 rows of the
rows of the  matrix for a cluster of size 4 with q = 2 log odds ratio parameters. The log odds ratio for the pairs (1 2), (1 3), (1 4) is
matrix for a cluster of size 4 with q = 2 log odds ratio parameters. The log odds ratio for the pairs (1 2), (1 3), (1 4) is 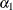 , and the log odds ratio for the pairs (2 3), (2 4), (3 4) is
, and the log odds ratio for the pairs (2 3), (2 4), (3 4) is 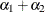 .
.
The quasi-likelihood information criterion (QIC) was developed by Pan (2001) as a modification of the Akaike information criterion (AIC) to apply to models fit by GEEs.
Define the quasi-likelihood under the independence working correlation assumption, evaluated with the parameter estimates under the working correlation of interest as
where the quasi-likelihood contribution of the jth observation in the ith cluster is defined in the section Quasi-likelihood Functions and ![]() are the parameter estimates obtained from GEEs with the working correlation of interest R.
are the parameter estimates obtained from GEEs with the working correlation of interest R.
QIC is defined as
where ![]() is the robust covariance estimate and
is the robust covariance estimate and ![]() is the inverse of the model-based covariance estimate under the independent working correlation assumption, evaluated at
is the inverse of the model-based covariance estimate under the independent working correlation assumption, evaluated at
![]() , the parameter estimates obtained from GEEs with the working correlation of interest R.
, the parameter estimates obtained from GEEs with the working correlation of interest R.
PROC GENMOD also computes an approximation to ![]() defined by Pan (2001) as
defined by Pan (2001) as
where p is the number of regression parameters.
Pan (2001) notes that QIC is appropriate for selecting regression models and working correlations, whereas ![]() is appropriate only for selecting regression models.
is appropriate only for selecting regression models.
See McCullagh and Nelder (1989) and Hardin and Hilbe (2003) for discussions of quasi-likelihood functions. The contribution of observation j in cluster i to the quasi-likelihood function evaluated at the regression parameters ![]() is given by
is given by ![]() , where
, where ![]() is defined in the following list. These are used in the computation of the quasi-likelihood information criteria (QIC) for
goodness of fit of models fit with GEEs. The
is defined in the following list. These are used in the computation of the quasi-likelihood information criteria (QIC) for
goodness of fit of models fit with GEEs. The ![]() are prior weights, if any, specified with the WEIGHT or FREQ statements. Note that the definition of the quasi-likelihood
for the negative binomial differs from that given in McCullagh and Nelder (1989). The definition used here allows the negative binomial quasi-likelihood to approach the Poisson as
are prior weights, if any, specified with the WEIGHT or FREQ statements. Note that the definition of the quasi-likelihood
for the negative binomial differs from that given in McCullagh and Nelder (1989). The definition used here allows the negative binomial quasi-likelihood to approach the Poisson as ![]() .
.
-
Normal:
![\[ Q_{ij} = -\frac{1}{2} w_{ij}(y_{ij}-\mu _{ij})^2 \]](images/statug_genmod0485.png)
-
Inverse Gaussian:
![\[ Q_{ij} = \frac{w_{ij}(\mu _{ij} - .5y_{ij})}{\mu _{ij}^2 } \]](images/statug_genmod0486.png)
-
Gamma:
![\[ Q_{ij} = -w_{ij}\left[ \frac{y_{ij}}{\mu _{ij}} + \log (\mu _{ij}) \right] \]](images/statug_genmod0487.png)
-
Negative binomial:
![\[ Q_{ij} = w_{ij}\left[ \log \Gamma \left( y_{ij} + \frac{1}{k}\right) - \log \Gamma \left(\frac{1}{k}\right) + y_{ij}\log \left(\frac{k\mu _{ij}}{1+k\mu _{ij}}\right) + \frac{1}{k}\log \left(\frac{1}{1+k\mu _{ij}}\right)\right] \]](images/statug_genmod0488.png)
-
Poisson:
![\[ Q_{ij} = w_{ij}(y_{ij} \log (\mu _{ij}) - \mu _{ij} ) \]](images/statug_genmod0489.png)
-
Binomial:
![\[ Q_{ij} = w_{ij}[r_{ij} \log (p_{ij}) + (n_{ij}-r_{ij}) \log (1-p_{ij})] \]](images/statug_genmod0490.png)
-
Multinomial (s categories):
![\[ Q_{ij} = w_{ij}\sum _{k=1}^ s y_{ijk}\log (\mu _{ijk}) \]](images/statug_genmod0491.png)
Boos (1992) and Rotnitzky and Jewell (1990) describe score tests applicable to testing ![]() in GEEs, where
in GEEs, where ![]() is a user-specified
is a user-specified ![]() contrast matrix or a contrast for a Type 3 test of hypothesis.
contrast matrix or a contrast for a Type 3 test of hypothesis.
Let ![]() be the regression parameters resulting from solving the GEE under the restricted model
be the regression parameters resulting from solving the GEE under the restricted model ![]() , and let
, and let ![]() be the generalized estimating equation values at
be the generalized estimating equation values at ![]() .
.
The generalized score statistic is
where ![]() is the model-based covariance estimate and
is the model-based covariance estimate and ![]() is the empirical covariance estimate. The p-values for T are computed based on the chi-square distribution with r degrees of freedom.
is the empirical covariance estimate. The p-values for T are computed based on the chi-square distribution with r degrees of freedom.