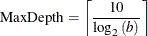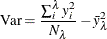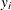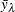-
ASSIGNMISSING=BRANCH |NONE |POPULAR |SIMILAR
-
specifies how PROC HPSPLIT creates a default splitting rule to handle missing values, unknown levels, and levels that have
fewer observations than you specify in the MINCATSIZE= option. An unknown level is a level of a categorical predictor that
does not exist in the training data but is encountered during scoring.
Both the ASSIGNMISSING= and NSURROGATES= options affect training and scoring. See the NSURROGATES= option for the definition
of surrogate rules.
During training, the primary splitting rule is created first, along with the default splitting rule (controlled by the ASSIGNMISSING=
option). If you request surrogate rules (by using the NSURROGATES= option), they are created after the primary and default
splits are made. When the splitting rules have been created, the rules are used as described in the following list for assigning
the training data by using the new splitting rules, and splitting rule creation continues on the new children.
Observation assignment during the training phase and during scoring proceeds as follows:
-
The primary splitting rule is applied if the rule’s variable is not missing. Otherwise:
-
The first surrogate rule (with the largest agreement, described in the section Primary and Surrogate Splitting Rules) is applied if the first surrogate rule’s variable is not missing. Otherwise:
-
Each subsequent (ordered by agreement) splitting rule is applied as described in the first surrogate item in this list. If
all of the surrogate rules’ variables are missing, then:
-
The default splitting rule is used.
Because there is always a default splitting rule, all data can be scored, even if the primary rule and all surrogate rules
cannot be used on a particular observation.
You can specify one of the following:
-
BRANCH
-
specifies that PROC HPSPLIT create a special child (branch) for the default rule and assign to that child missing values,
unknown levels, and levels that have fewer observations than you specify in the MINCATSIZE= option. You need to have missing
values in the training data set for this to work. If no missing values or levels that have fewer observations than you specify
in the MINCATSIZE= option are available at the split in the training data set, then missing values are assigned using ASSIGNMISSING=POPULAR.
-
NONE
-
specifies that observations that have any missing variables be excluded from the analysis. In addition, the default rule (to
handle unknown levels, which include missing values, because they are excluded from the analysis, and levels that have fewer
observations than you specify in the MINCATSIZE= option) is created using ASSIGNMISSING=POPULAR.
-
POPULAR
-
specifies that missing values be assigned to the most popular (largest) child.
-
SIMILAR
-
specifies that missing values be assigned to the child that they are most similar to (using the chi-square for categorical
responses or F-test criterion for continuous responses). For more information about this option and the similarity measurement, see the
section Handling Missing Values. If no missing values or levels that have fewer observations than you specify in the MINCATSIZE= option are available at
the split in the training data set, then missing values are assigned using ASSINGMISSING=POPULAR.
By default, ASSIGNMISSING=NONE.
-
CVCC
CVCOSTCOMPLEXITY
-
requests a table of the results of cost-complexity pruning based on cross validation. For each fold in the cross validation,
the table provides the penalty parameter, the number of leaves, and the average ASE. In addition, the table provides the minimum
and maximum ASE, and the minimum, median, and maximum number of leaves. The number of leaves is the floor of the median. You
can use the PLOTS=CVCC option to request a plot of the information in this table.
-
CVMETHOD=NONE |RANDOM <(k)>
-
requests the cross validation method to be performed.
You can specify one of the following:
-
NONE
-
suppresses cross validation.
-
RANDOM <(k)>
-
assigns each training observation randomly to one of the k folds (with a probability of 1/k for any given fold) for cross validation. The default value of k is 10.
Cross validation applies to the CVMODELFIT option and cost-complexity pruning. In k-fold cross validation, the training set is divided into k folds; k trees are then built using all but the one fold. For example, the first tree uses all of the training set except for the
observations in the first fold. The second uses all of the training set except for the observations in the second fold, and
so on.
The holdout fold for each tree is used to calculate the ASE of that tree. The average ASE across the k trees is the cross validation error for that set of trees. This process is repeated for each value of the parameter that
is cross validated. The parameter that has the minimum cross validated error is used as the best parameter value. A final
tree is then grown using this final parameter value.
Note: Attempting to use a PARTITION
statement along with cross validation results in an error.
By default, CVMETHOD=RANDOM(10) when you perform cost-complexity pruning with no PARTITION
statement.
-
CVMODELFIT
-
requests model assessment with cross validation. When you specify this option, the procedure does a cross validation of the
final model parameters and produces a table that describes the cross validation error measures of the parameters. The table
contains various summary statistics of the ASE, the number of leaves, and, for a categorical response, the misclassification
rate across the k trees grown. This option also requests a table that contains the cross validation confusion matrix.
If cost complexity is cross validated (the default if you use cost-complexity pruning without a validation set), the assessment
is a completely separate cross validation of the final tree penalty parameter, using a different seed for fold assignment.
Note: The assessment is not on a holdout sample but instead on k-fold cross validation. This is not a second run of the procedure but instead is done automatically and internally.
-
DATA=SAS-data-set
-
names the predictor SAS data set to be used by PROC HPSPLIT. The default is the most recently created data set.
If the procedure executes in distributed mode, the predictor data are distributed to memory on the appliance nodes and analyzed
in parallel, unless the data are already distributed in the appliance database. In that case, the procedure reads the data
alongside the distributed database. For more information, see the section Processing Modes about the various execution modes and the section Alongside-the-Database Execution about the alongside-the-database model.
-
INTERVALBINS=number
-
specifies the number of bins for continuous variables. PROC HPSPLIT bins continuous predictors to a fixed bin size. This option controls the number
of bins and thereby also the size of the bins. For more information about interval variable binning, see the section Details: HPSPLIT Procedure.
By default, INTERVALBINS=100.
-
LEVTHRESH1=number
-
applies only to categorical predictor variables and specifies the limit for the number of computations in an exhaustive search
for the optimal partition of the levels of a particular variable. The splitter first evaluates the number of computations
that are needed for an exhaustive search. If this number exceeds the limit, then the splitter falls back to a faster heuristic
algorithm. You can use the LEVTHRESH2= option to specify the limit for the number of computations in this faster algorithm.
By default, LEVTHRESH1=500000.
-
LEVTHRESH2=number
-
applies to categorical predictor variables and continuous predictor variables with multiway splits. This option does not apply
to continuous predictor variables with binary splits.
For a categorical predictor variable, the splitter first evaluates the number of computations that are needed for an exhaustive
search. If this number exceeds the limit that you specify in the LEVTHRESH1= option, then the splitter falls back to a faster
heuristic algorithm. You can use the LEVTHRESH2= option to specify the limit for the number of computations in this faster
algorithm. If this number of computations exceeds the limit that you specify in the LEVTHRESH2= option, then the splitter
falls back to an even faster algorithm.
For a continuous predictor variable, the splitter first tries to perform an exhaustive search for the optimal split values.
The splitter first evaluates the number of computations that are needed for an exhaustive search. If this number exceeds the
limit that you specify in the LEVTHRESH2= option, the splitter falls back to a faster heuristic algorithm.
By default, LEVTHRESH2=1000000.
-
MAXBRANCH=b
-
specifies the maximum number of children per node in the tree. PROC HPSPLIT tries to create this number of children unless
it is impossible (for example, if a split variable does not have enough levels). By default, MAXBRANCH=2.
-
MAXDEPTH=number
-
specifies the maximum depth of the tree to be grown. The default is set using the following equation, where b is the value for the MAXBRANCH=
option:
-
MINCATSIZE=number
-
specifies the number of observations that a categorical variable level must have in order to be considered in the split. Predictor variable levels
that have fewer observations than number receive the nonsurrogate missing value assignment for that split. See the NSURROGATES= option for the definition of surrogate
rules.
By default, MINCATSIZE=1.
-
MINLEAFSIZE=number
-
specifies the minimum number of observations that each child of a split must contain in the training data set in order for the split to be considered.
By default, MINLEAFSIZE=1.
-
MINVARIANCE=value
-
specifies the minimum variance for a regression tree leaf to be eligible for splitting. That is, leaves whose variance is
less than value are not split any further.
By default, MINVARIANCE=1E–8.
The variance at some leaf 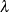 with weight
with weight 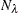 (number of observations) is calculated using
(number of observations) is calculated using
where 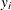 is the response value at observation i and
is the response value at observation i and 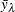 is the average value of the response within leaf
is the average value of the response within leaf 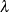 .
.
-
NODES=DETAIL |SUMMARY
-
requests a table that contains the description of the paths from each leaf to the root.
You can specify the following values:
-
DETAIL
-
prints all nodes and leaves, including the path from each node or leaf to the root of the tree.
-
SUMMARY
-
prints all nodes and leaves, without including the path to the root of the tree.
-
NOPRINT
-
suppresses the generation of ODS output.
-
NSURROGATES=number
-
specifies the number of surrogate rules to create for each splitting rule. Surrogate rules are backup splitting rules that are used when the variable
that corresponds to the primary splitting rule is missing. By default, NSURROGATES=0.
Both the ASSIGNMISSING= option and NSURROGATES= options affect training and scoring.
During training, the primary splitting rule is created first, along with the default splitting rule (controlled by the ASSIGNMISSING=
option). If you request surrogate rules (by specifying the NSURROGATES= option), they are created after the primary and default
splits are created. When the splitting rules have been created, the rules are used as described in the following list for
assigning the training data by using the new splitting rules, and splitting rule creation continues on the new children.
Observation assignment during the training phase and during scoring proceeds as follows:
-
The primary splitting rule is applied if the primary rule’s variable is not missing. Otherwise:
-
The first surrogate rule (with the largest agreement, described in the section Primary and Surrogate Splitting Rules) is applied if the first surrogate rule’s variable is not missing. Otherwise:
-
Each subsequent (ordered by agreement) splitting rule is applied as described in the first surrogate item in the list. If
all of the surrogate rules’ variables are missing, then:
-
The default splitting rule is used.
Because there is always a default splitting rule, all data can be scored, even if the primary rule and all surrogate rules
cannot be used on a particular observation.
As auxiliary rules, surrogate rules are used in the following ways:
-
Surrogate rules are used in generating the tree.
-
Surrogate rules therefore also affect the final tree metrics.
-
Surrogate rules are used in the SAS code output from the CODE
statement.
-
Surrogate rules are used in scoring the predictor data set by using the OUTPUT
statement.
-
Surrogate rules are not written out in the “Node Rules” file, output from the RULES
statement.
-
PLOTS <(global-plot-option)> <= plot-request <(options)>>
PLOTS <(global-plot-option)> <= (plot-request <(options)> <... plot-request <(options)>>)>
-
controls the plots that are produced through ODS Graphics. When you specify only one plot-request, you can omit the parentheses around it. Some examples follow.
You can specify the following global-plot-option:
-
ONLY
-
suppresses the default plots. Only plots that you specifically request are displayed.
You can specify the following plot-requests:
-
ALL
-
produces all appropriate plots.
-
CVCC
-
produces the cross validation plot. This option is enabled by default if you request cross validation of cost-complexity pruning.
-
NONE
-
suppresses the default plots. Only plots that you specifically request are displayed.
-
PRUNEUNTIL
-
produces a plot of the metric that is used to select the final subtree. This option is enabled by default except in the following
cases:
-
You request cross validation of cost-complexity pruning.
-
You specify the LEAVES= option in the PRUNE
statement when you use the cost-complexity pruning method.
-
You specify the OFF option in the PRUNE
statement. Note that specifying the PRUNEUNTIL option has no effect in this case.
-
ROC
-
produces the receiver operating characteristic (ROC) curve. This option is enabled by default.
-
WHOLETREE <(whole-tree-options)>
-
produces a plot to visualize the entire finished (grown and pruned) tree. This option is enabled by default.
You can specify the following whole-tree-options:
-
LINKSTYLE=link-style
-
specifies the style of links between nodes and leaves in the tree. You can specify the following link-styles:
-
CURVED
-
requests curved links between the nodes and their children.
-
ORTHOGONAL
-
requests that links go straight down partway from a node to its children, create a horizontal line at the base of the vertical
line, and then go straight down from that line to each child.
-
STRAIGHT
-
requests that links go straight from the nodes to their children.
By default, LINKSTYLE=CURVED.
-
LINKWIDTH=link-width
-
specifies the width of links between nodes and leaves in the tree. You can specify the following link-widths:
-
CONSTANT
-
requests that all links have the same thickness.
-
PROPORTIONAL
-
requests that links have a thickness proportional to the total number of observations that go between the node and each child.
By default, LINKWIDTH=PROPORTIONAL.
-
NOLEGEND
-
turns off the legend.
-
ZOOMEDTREE <(zoomed-tree-options)>
-
produces a plot to visualize a portion of the finished (grown and pruned) tree. This option is enabled by default.
You can specify the following zoomed-tree-options:
-
DEPTH=depth
-
requests that PROC HPSPLIT create plots specified by the ZOOMEDTREE option for each node-id down to depth.
-
LINKSTYLE=link-style
-
specifies the style of links between nodes and leaves in the tree. You can specify the following link-styles:
-
CURVED
-
requests curved links between the nodes and their children.
-
ORTHOGONAL
-
requests that links go straight down partway from a node to its children, create a horizontal line at the base of the vertical
line, and then go straight down from that line to each child.
-
STRAIGHT
-
requests that links go straight from the nodes to their children.
By default, LINKSTYLE=CURVED.
-
LINKWIDTH=link-width
-
specifies the width of links between nodes and leaves in the tree. You can specify the following link-widths:
-
CONSTANT
-
requests that all links have the same thickness.
-
PROPORTIONAL
-
requests that links have a thickness proportional to the total number of observations that go between the node and each child.
By default, LINKWIDTH=PROPORTIONAL.
-
NODES=(node-id <node-id <...>>)
-
requests plots for the subtree that is rooted at nodes at node-id. The default node ID is “0,” the root of the entire tree. The values of node-id are alphanumeric strings that are displayed within the nodes in the plot that is created by the WHOLETREE option. PROC HPSPLIT
creates one plot specified by the ZOOMEDTREE option for each node-id that you specify.
-
NOLEGEND
-
turns off the legend
-
SEED=number
-
specifies the initial seed for random number generation for cross validation. The value of number must be an integer. The default seed is based on the date and time.
-
SPLITONCE
-
specifies that variables be split only once on a branch. However, a variable can be used more than once across branches. That
is, a variable cannot be split more than once on the path from any leaf to the root node.