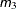PARMS <(value-list)…> </ options> ;
The PARMS statement specifies initial values for the covariance parameters, or it requests a grid search over several values
of these parameters. You must specify the values in the order in which they appear in the “Covariance Parameter Estimates” table.
The value-list specification can take any of several forms:
- m
-
a single value
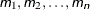
-
several values
- m to n
-
a sequence in which m equals the starting value, n equals the ending value, and the increment equals 1
- m to n by i
-
a sequence in which m equals the starting value, n equals the ending value, and the increment equals i
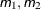 to
to 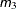
-
mixed values and sequences
You can use the PARMS statement to input known parameters.
If you specify more than one set of initial values, PROC HPLMIXED performs a grid search of the likelihood surface and uses
the best point on the grid for subsequent analysis. Specifying a large number of grid points can result in long computing
times.
The results from the PARMS statement are the values of the parameters on the specified grid (denoted by CovP1 through CovPn), the residual variance (possibly estimated) for models with a residual variance parameter, and various functions of the
likelihood.
You can specify the following options in the PARMS statement after a slash (/).
-
HOLD=all
EQCONS=all
-
specifies that all parameter values be held to equal the specified values.
For example, the following statement constrains all covariance parameters to equal 5, 3, 2, and 3:
parms (5) (3) (2) (3) / hold=all;
-
LOWERB=value-list
-
enables you to specify lower boundary constraints on the covariance parameters. The value-list specification is a list of numbers or missing values (.) separated by commas. You must list the numbers in the order that
PROC HPLMIXED uses for the covariance parameters, and each number corresponds to the lower boundary constraint. A missing
value instructs PROC HPLMIXED to use its default constraint. If you do not specify numbers for all of the covariance parameters,
PROC HPLMIXED assumes the remaining ones are missing.
This option is useful when you want to constrain the 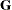 matrix to be positive definite in order to avoid the more computationally intensive algorithms that would be required when
matrix to be positive definite in order to avoid the more computationally intensive algorithms that would be required when
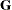 becomes singular. The corresponding statements for a random coefficients model are as follows:
becomes singular. The corresponding statements for a random coefficients model are as follows:
proc hplmixed;
class person;
model y = time;
random int time / type=fa0(2) sub=person;
parms / lowerb=1e-4,.,1e-4;
run;
The TYPE=FA0(2) structure specifies a Cholesky root parameterization for the 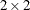 unstructured blocks in
unstructured blocks in  . This parameterization ensures that the
. This parameterization ensures that the 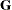 matrix is nonnegative definite, and the PARMS statement then ensures that it is positive definite by constraining the two
diagonal terms to be greater than or equal to 1E–4.
matrix is nonnegative definite, and the PARMS statement then ensures that it is positive definite by constraining the two
diagonal terms to be greater than or equal to 1E–4.
-
NOITER
-
requests that no optimization iterations be performed and that PROC HPLMIXED use the best value from the grid search to perform
inferences. By default, iterations begin at the best value from the PARMS grid search. The NOITER option will be implied by
the specification of the BLUP option in the HPLMIXED statement.
-
PARMSDATA=SAS-data-set
PDATA=SAS-data-set
-
reads in covariance parameter values from a SAS data set. The data set should contain the Est or Covp1 through Covpn variables.
-
UPPERB=value-list
-
enables you to specify upper boundary constraints on the covariance parameters. The value-list specification is a list of numbers or missing values (.) separated by commas. You must list the numbers in the order that
PROC HPLMIXED uses for the covariance parameters, and each number corresponds to the upper boundary constraint. A missing
value instructs PROC HPLMIXED to use its default constraint. If you do not specify numbers for all of the covariance parameters,
PROC HPLMIXED assumes that the remaining ones are missing.
Copyright © SAS Institute Inc. All Rights Reserved.
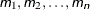
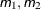 to
to