The UNIVARIATE Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Missing ValuesRoundingDescriptive StatisticsCalculating the ModeCalculating PercentilesTests for LocationConfidence Limits for Parameters of the Normal DistributionRobust EstimatorsCreating Line Printer PlotsCreating High-Resolution GraphicsUsing the CLASS Statement to Create Comparative PlotsPositioning InsetsFormulas for Fitted Continuous DistributionsGoodness-of-Fit TestsKernel Density EstimatesConstruction of Quantile-Quantile and Probability PlotsInterpretation of Quantile-Quantile and Probability PlotsDistributions for Probability and Q-Q PlotsEstimating Shape Parameters Using Q-Q PlotsEstimating Location and Scale Parameters Using Q-Q PlotsEstimating Percentiles Using Q-Q PlotsInput Data SetsOUT= Output Data Set in the OUTPUT StatementOUTHISTOGRAM= Output Data SetOUTKERNEL= Output Data SetOUTTABLE= Output Data SetTables for Summary StatisticsODS Table NamesODS Tables for Fitted DistributionsODS GraphicsComputational Resources
Missing ValuesRoundingDescriptive StatisticsCalculating the ModeCalculating PercentilesTests for LocationConfidence Limits for Parameters of the Normal DistributionRobust EstimatorsCreating Line Printer PlotsCreating High-Resolution GraphicsUsing the CLASS Statement to Create Comparative PlotsPositioning InsetsFormulas for Fitted Continuous DistributionsGoodness-of-Fit TestsKernel Density EstimatesConstruction of Quantile-Quantile and Probability PlotsInterpretation of Quantile-Quantile and Probability PlotsDistributions for Probability and Q-Q PlotsEstimating Shape Parameters Using Q-Q PlotsEstimating Location and Scale Parameters Using Q-Q PlotsEstimating Percentiles Using Q-Q PlotsInput Data SetsOUT= Output Data Set in the OUTPUT StatementOUTHISTOGRAM= Output Data SetOUTKERNEL= Output Data SetOUTTABLE= Output Data SetTables for Summary StatisticsODS Table NamesODS Tables for Fitted DistributionsODS GraphicsComputational Resources -
Examples
 Computing Descriptive Statistics for Multiple VariablesCalculating ModesIdentifying Extreme Observations and Extreme ValuesCreating a Frequency TableCreating Basic Summary PlotsAnalyzing a Data Set With a FREQ VariableSaving Summary Statistics in an OUT= Output Data SetSaving Percentiles in an Output Data SetComputing Confidence Limits for the Mean, Standard Deviation, and VarianceComputing Confidence Limits for Quantiles and PercentilesComputing Robust EstimatesTesting for LocationPerforming a Sign Test Using Paired DataCreating a HistogramCreating a One-Way Comparative HistogramCreating a Two-Way Comparative HistogramAdding Insets with Descriptive StatisticsBinning a HistogramAdding a Normal Curve to a HistogramAdding Fitted Normal Curves to a Comparative HistogramFitting a Beta CurveFitting Lognormal, Weibull, and Gamma CurvesComputing Kernel Density EstimatesFitting a Three-Parameter Lognormal CurveAnnotating a Folded Normal CurveCreating Lognormal Probability PlotsCreating a Histogram to Display Lognormal FitCreating a Normal Quantile PlotAdding a Distribution Reference LineInterpreting a Normal Quantile PlotEstimating Three Parameters from Lognormal Quantile PlotsEstimating Percentiles from Lognormal Quantile PlotsEstimating Parameters from Lognormal Quantile PlotsComparing Weibull Quantile PlotsCreating a Cumulative Distribution PlotCreating a P-P Plot
Computing Descriptive Statistics for Multiple VariablesCalculating ModesIdentifying Extreme Observations and Extreme ValuesCreating a Frequency TableCreating Basic Summary PlotsAnalyzing a Data Set With a FREQ VariableSaving Summary Statistics in an OUT= Output Data SetSaving Percentiles in an Output Data SetComputing Confidence Limits for the Mean, Standard Deviation, and VarianceComputing Confidence Limits for Quantiles and PercentilesComputing Robust EstimatesTesting for LocationPerforming a Sign Test Using Paired DataCreating a HistogramCreating a One-Way Comparative HistogramCreating a Two-Way Comparative HistogramAdding Insets with Descriptive StatisticsBinning a HistogramAdding a Normal Curve to a HistogramAdding Fitted Normal Curves to a Comparative HistogramFitting a Beta CurveFitting Lognormal, Weibull, and Gamma CurvesComputing Kernel Density EstimatesFitting a Three-Parameter Lognormal CurveAnnotating a Folded Normal CurveCreating Lognormal Probability PlotsCreating a Histogram to Display Lognormal FitCreating a Normal Quantile PlotAdding a Distribution Reference LineInterpreting a Normal Quantile PlotEstimating Three Parameters from Lognormal Quantile PlotsEstimating Percentiles from Lognormal Quantile PlotsEstimating Parameters from Lognormal Quantile PlotsComparing Weibull Quantile PlotsCreating a Cumulative Distribution PlotCreating a P-P Plot - References
This example shows how to display a fitted curve that is not supported by the HISTOGRAM statement. The offset of an attachment
point is measured (in mm) for a number of manufactured assemblies, and the measurements (Offset) are saved in a data set named Assembly. The following statements create the data set Assembly:
data Assembly; label Offset = 'Offset (in mm)'; input Offset @@; datalines; 11.11 13.07 11.42 3.92 11.08 5.40 11.22 14.69 6.27 9.76 9.18 5.07 3.51 16.65 14.10 9.69 16.61 5.67 2.89 8.13 9.97 3.28 13.03 13.78 3.13 9.53 4.58 7.94 13.51 11.43 11.98 3.90 7.67 4.32 12.69 6.17 11.48 2.82 20.42 1.01 3.18 6.02 6.63 1.72 2.42 11.32 16.49 1.22 9.13 3.34 1.29 1.70 0.65 2.62 2.04 11.08 18.85 11.94 8.34 2.07 0.31 8.91 13.62 14.94 4.83 16.84 7.09 3.37 0.49 15.19 5.16 4.14 1.92 12.70 1.97 2.10 9.38 3.18 4.18 7.22 15.84 10.85 2.35 1.93 9.19 1.39 11.40 12.20 16.07 9.23 0.05 2.15 1.95 4.39 0.48 10.16 4.81 8.28 5.68 22.81 0.23 0.38 12.71 0.06 10.11 18.38 5.53 9.36 9.32 3.63 12.93 10.39 2.05 15.49 8.12 9.52 7.77 10.70 6.37 1.91 8.60 22.22 1.74 5.84 12.90 13.06 5.08 2.09 6.41 1.40 15.60 2.36 3.97 6.17 0.62 8.56 9.36 10.19 7.16 2.37 12.91 0.95 0.89 3.82 7.86 5.33 12.92 2.64 7.92 14.06 ;
It is decided to fit a folded normal distribution to the offset measurements. A variable X has a folded normal distribution if ![]() , where Y is distributed as
, where Y is distributed as ![]() . The fitted density is
. The fitted density is
where ![]() .
.
You can use SAS/IML to compute preliminary estimates of ![]() and
and ![]() based on a method of moments given by Elandt (1961). These estimates are computed by solving equation (19) Elandt (1961), which is given by
based on a method of moments given by Elandt (1961). These estimates are computed by solving equation (19) Elandt (1961), which is given by
where ![]() is the standard normal distribution function and
is the standard normal distribution function and
Then the estimates of ![]() and
and ![]() are given by
are given by
![\[ \begin{array}{lll} \hat{\sigma }_0 & = \sqrt { \frac{ \frac{1}{n} \sum _{i=1}^ n x_ i^2}{1 + \hat{\theta }^2 } } \\ \hat{mu}_0 & = \hat{\theta } \cdot \hat{\sigma }_0 \end{array} \]](images/procstat_univariate0509.png)
Begin by using PROC MEANS to compute the first and second moments and by using the following DATA step to compute the constant A:
proc means data = Assembly noprint; var Offset; output out=stat mean=m1 var=var n=n min = min; run; * Compute constant A from equation (19) of Elandt (1961); data stat; keep m2 a min; set stat; a = (m1*m1); m2 = ((n-1)/n)*var + a; a = a/m2; run;
Next, use the SAS/IML subroutine NLPDD to solve equation (19) by minimizing ![]() , and compute
, and compute ![]() and
and ![]() :
:
proc iml;
use stat;
read all var {m2} into m2;
read all var {a} into a;
read all var {min} into min;
* f(t) is the function in equation (19) of Elandt (1961);
start f(t) global(a);
y = .39894*exp(-0.5*t*t);
y = (2*y-(t*(1-2*probnorm(t))))**2/(1+t*t);
y = (y-a)**2;
return(y);
finish;
* Minimize (f(t)-A)**2 and estimate mu and sigma;
if ( min < 0 ) then do;
print "Warning: Observations are not all nonnegative.";
print " The folded normal is inappropriate.";
stop;
end;
if ( a < 0.637 ) then do;
print "Warning: the folded normal may be inappropriate";
end;
opt = { 0 0 };
con = { 1e-6 };
x0 = { 2.0 };
tc = { . . . . . 1e-8 . . . . . . .};
call nlpdd(rc,etheta0,"f",x0,opt,con,tc);
esig0 = sqrt(m2/(1+etheta0*etheta0));
emu0 = etheta0*esig0;
create prelim var {emu0 esig0 etheta0};
append;
close prelim;
* Define the log likelihood of the folded normal;
start g(p) global(x);
y = 0.0;
do i = 1 to nrow(x);
z = exp( (-0.5/p[2])*(x[i]-p[1])*(x[i]-p[1]) );
z = z + exp( (-0.5/p[2])*(x[i]+p[1])*(x[i]+p[1]) );
y = y + log(z);
end;
y = y - nrow(x)*log( sqrt( p[2] ) );
return(y);
finish;
* Maximize the log likelihood with subroutine NLPDD;
use assembly;
read all var {offset} into x;
esig0sq = esig0*esig0;
x0 = emu0||esig0sq;
opt = { 1 0 };
con = { . 0.0, . . };
call nlpdd(rc,xr,"g",x0,opt,con);
emu = xr[1];
esig = sqrt(xr[2]);
etheta = emu/esig;
create parmest var{emu esig etheta};
append;
close parmest;
quit;
The preliminary estimates are saved in the data set Prelim, as shown in Output 4.25.1.
Now, using ![]() and
and ![]() as initial estimates, call the NLPDD subroutine to maximize the log likelihood,
as initial estimates, call the NLPDD subroutine to maximize the log likelihood, ![]() , of the folded normal distribution, where, up to a constant,
, of the folded normal distribution, where, up to a constant,
* Define the log likelihood of the folded normal;
start g(p) global(x);
y = 0.0;
do i = 1 to nrow(x);
z = exp( (-0.5/p[2])*(x[i]-p[1])*(x[i]-p[1]) );
z = z + exp( (-0.5/p[2])*(x[i]+p[1])*(x[i]+p[1]) );
y = y + log(z);
end;
y = y - nrow(x)*log( sqrt( p[2] ) );
return(y);
finish;
* Maximize the log likelihood with subroutine NLPDD;
use assembly;
read all var {offset} into x;
esig0sq = esig0*esig0;
x0 = emu0||esig0sq;
opt = { 1 0 };
con = { . 0.0, . . };
call nlpdd(rc,xr,"g",x0,opt,con);
emu = xr[1];
esig = sqrt(xr[2]);
etheta = emu/esig;
create parmest var{emu esig etheta};
append;
close parmest;
quit;
The data set ParmEst contains the maximum likelihood estimates ![]() and
and ![]() (as well as
(as well as ![]() ), as shown in Output 4.25.2.
), as shown in Output 4.25.2.
To annotate the curve on a histogram, begin by computing the width and endpoints of the histogram intervals. The following
statements save these values in a data set called OutCalc. Note that a plot is not produced at this point.
proc univariate data = Assembly noprint;
histogram Offset / outhistogram = out normal(noprint) noplot;
run;
data OutCalc (drop = _MIDPT_);
set out (keep = _MIDPT_) end = eof;
retain _MIDPT1_ _WIDTH_;
if _N_ = 1 then _MIDPT1_ = _MIDPT_;
if eof then do;
_MIDPTN_ = _MIDPT_;
_WIDTH_ = (_MIDPTN_ - _MIDPT1_) / (_N_ - 1);
output;
end;
run;
Output 4.25.3 provides a listing of the data set OutCalc. The width of the histogram bars is saved as the value of the variable _WIDTH_; the midpoints of the first and last histogram bars are saved as the values of the variables _MIDPT1_ and _MIDPTN_.
The following statements create an annotate data set named Anno, which contains the coordinates of the fitted curve:
data Anno;
merge ParmEst OutCalc;
length function color $ 8;
function = 'point';
color = 'black';
size = 2;
xsys = '2';
ysys = '2';
when = 'a';
constant = 39.894*_width_;;
left = _midpt1_ - .5*_width_;
right = _midptn_ + .5*_width_;
inc = (right-left)/100;
do x = left to right by inc;
z1 = (x-emu)/esig;
z2 = (x+emu)/esig;
y = (constant/esig)*(exp(-0.5*z1*z1)+exp(-0.5*z2*z2));
output;
function = 'draw';
end;
run;
The following statements read the ANNOTATE= data set and display the histogram and fitted curve:
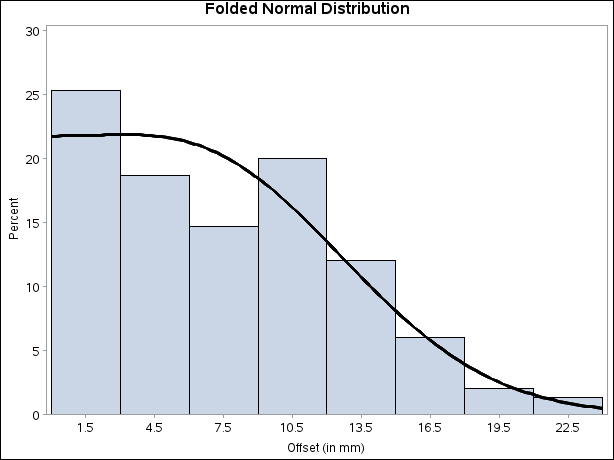
title 'Folded Normal Distribution'; ods graphics off; proc univariate data=assembly noprint; histogram Offset / annotate = anno; run;
Output 4.25.4 displays the histogram and fitted curve.
A sample program for this example, uniex15.sas, is available in the SAS Sample Library for Base SAS software.