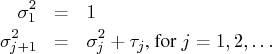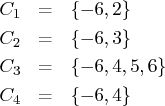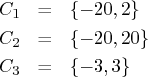| Language Reference |
SEQ, SEQSCALE, and SEQSHIFT Calls
perform discrete sequential tests
- CALL SEQ( prob, domain <, <TSCALE=tscale<, <EPS=eps>
- <, <DEN=den>);
- CALL SEQSCALE( prob, gscale, domain, level<, <IGUESS=iguess>
- <, <TSCALE=tscale<, <EPS=eps<, <DEN=den>);
- CALL SEQSHIFT( prob, shift, domain, plevel<, <IGUESS=iguess>
- <, <TSCALE=tscale<, <EPS=eps<, <DEN=den>);
The SEQSHIFT subroutine returns the following values:
- prob
- is an
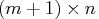 matrix.
The
matrix.
The ![[i,j]](images/langref_langrefeq1160.gif) entry in the array contains the probability
at the
entry in the array contains the probability
at the ![[i,j]](images/langref_langrefeq1160.gif) entry of the argument domain.
Also, the probability at infinity at every level
entry of the argument domain.
Also, the probability at infinity at every level 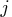 is
returned in the last entry (
is
returned in the last entry (![[m+1,j]](images/langref_langrefeq1161.gif) ) of column
) of column 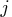 .
Upon a successful completion of any
routine, this variable is always returned.
.
Upon a successful completion of any
routine, this variable is always returned.
- gscale
- is a numeric variable that returns from the
routine SEQSCALE and contains the scaling of the
current geometry defined by domain that
would yield a given significance level level.
- shift
- is a numeric variable that returns from the routine SEQSHIFT and contains the shift of current geometry defined by domain that would yield a given power level plevel.
The inputs to the SEQSHIFT subroutine are as follows:
- domain
- specifies an
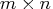 matrix containing
the boundary points separating the intervals of
continuation/stopping of the sequential test.
Each column
matrix containing
the boundary points separating the intervals of
continuation/stopping of the sequential test.
Each column 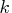 contains the boundary points at level
contains the boundary points at level 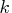 sorted in an ascending order, with .M and .P
representing
sorted in an ascending order, with .M and .P
representing 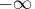 and
and 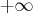 , respectively.
They must start on the first row, and any remaining
entries must be filled with a missing value.
Elements that follow the missing
value in any column are ignored.
The number of columns
, respectively.
They must start on the first row, and any remaining
entries must be filled with a missing value.
Elements that follow the missing
value in any column are ignored.
The number of columns 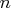 is equal to the
number of stages present in the sequential test.
The row dimension
is equal to the
number of stages present in the sequential test.
The row dimension 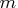 must be even, and it is equal
to the maximum number of boundary points in a level.
In fact, domain is the tabular
form of the finite boundary points.
Entries in domain with absolute values that
exceed a standardized value of 8 at any level
are internally reset to a standardized value
of 8 or -8, depending on the sign of the entry.
This is reflected in the results returned
for the probabilities and the densities.
must be even, and it is equal
to the maximum number of boundary points in a level.
In fact, domain is the tabular
form of the finite boundary points.
Entries in domain with absolute values that
exceed a standardized value of 8 at any level
are internally reset to a standardized value
of 8 or -8, depending on the sign of the entry.
This is reflected in the results returned
for the probabilities and the densities.
- tscale
- specifies an optional
 vector that describes
the time intervals between two consecutive stages.
In the absence of tscale, these time
intervals are internally set to 1.
The IML keyword for tscale is TSCALE.
vector that describes
the time intervals between two consecutive stages.
In the absence of tscale, these time
intervals are internally set to 1.
The IML keyword for tscale is TSCALE.
- eps
- specifies an optional numeric parameter for
controlling the absolute precision of the computation.
In the absence of eps, the precision
is internally set to 1E-7.
The IML keyword for eps is EPS.
- den
- specifies an optional character string to
describe the name of an
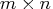 matrix.
The
matrix.
The ![[i,j]](images/langref_langrefeq1160.gif) entry in the matrix returns the density
of the distribution at the
entry in the matrix returns the density
of the distribution at the ![[i,j]](images/langref_langrefeq1160.gif) entry of the
matrix specified by the domain argument.
The IML keyword for den is DEN.
entry of the
matrix specified by the domain argument.
The IML keyword for den is DEN.
- iguess
- specifies an optional numeric parameter that contains an initial
guess for the variable gscale in the SEQSCALE subroutine
or for the variable mean in the SEQSHIFT subroutine.
In general, very good estimates for these initial guesses
can be provided by an iterative process, and these
estimates become extremely valuable near convergence.
The IML keyword for iguess is IGUESS.
- level
- specifies a numeric parameter in the SEQSCALE subroutine that
contains the required significance level to be achieved through
scaling the domain (see the description of SEQSCALE).
- plevel
- specifies a numeric parameter in the SEQSHIFT subroutine that provides the required power level to be achieved through shifting the domain (see the description of SEQSHIFT).
SEQ Call
To compute the probability from a sequential test, you must specify a matrix containing the boundaries. With the optional additional information concerning the time intervals and the target accuracy, or their default values, the SEQ subroutine returns the matrix that contains the probability and optionally returns the density from a sequential test evaluated at each given point of the boundary. Let
The SEQ call computes, with
SEQSCALE Call
Given a domain
For a significance level of
SEQSHIFT Call
Given a geometry
Actually, a simple transformation of the variables in the sequential problem yields the following result:
Many options are available with the NLP family of optimization routines, which are described in Chapter 4, "Nonlinear Optimization Subroutines."
Consider the following continuation intervals:
/* function to insert in m the geometry column a at level k*/
start table(m,a,k);
if ncol(m) = 0 & nrow(m) = 0 then m = j(nrow(a),k,.);
if nrow(m) < nrow(a) then m = m// j(nrow(a)-nrow(m),ncol(m),.);
if ncol(m) < k then m = m || j(nrow(m),k-ncol(m),.);
m[1:nrow(a),k] = a;
finish;
call table(m,{-6,2},1);
call table(m,{-6,3},2);
call table(m,{-6,4,5,6},3);
call table(m,{-6,4},4);
call seq(prob,m) eps = 1.e-8 den="density";
print m;
print prob;
print density;
The following output displays the values returned
for The probability at the level
Consider the continuation intervals
reset nocenter;
/* function to insert in m the geometry column a at level k*/
start table(m,a,k);
if ncol(m) = 0 & nrow(m) = 0 then m = j(nrow(a),k,.);
if nrow(m) < nrow(a) then m = m// j(nrow(a)-nrow(m),ncol(m),.);
if ncol(m) < k then m = m || j(nrow(m),k-ncol(m),.);
m[1:nrow(a),k] = a;
finish;
call table(m,{-20,2},1);
call table(m,{-20,20},2);
call table(m,{-3,3},3);
/**************************************/
/* TSCALE has the default value of 1 */
/**************************************/
call seq(prob1,m) eps = 1.e-8 den="density";
print m[format=f5.] prob1[format=e12.5];
call table(mm,{-20,2},1);
call table(mm,{-3,3},2);
/* You can show a 2-step separation between the levels */
/* while dropping the intermediate level at 2 */
tscale = { 2 };
call seq(prob2,mm) eps = 1.e-8 den="density" TSCALE=tscale;
print mm[format=f5.] prob2[format=e12.5];
The values returned for the variables
Some internal limitations are imposed on the geometry. Consider the three-level case with geometry
The computations performed by the first call of the SEQ subroutine can be simplified since the second level is large enough to be considered infinite. The matrix MM contains the first and third columns of the matrix M. However, in order to specify the two-step separation between the levels, you must specify tscale=2.
This example verifies some of the results published in Table 3 of Pocock (1982). That is, the following IML program verifies for the given domain that the significance level is 0.05 and that the power is
/*****************************************/
/* first check whether the numbers yield */
/* 0.95 for the alpha level */
/*****************************************/
bm ={-3.663 -2.884 -2.573 -2.375 -2.037,
-2.988 -2.537 -2.407 -2.346 -2.156,
-2.598 -2.390 -2.390 -2.390 -2.310,
-2.446 -2.404 -2.404 -2.404 -2.396};
bplevel = { 0.5 0.25 0.1 0.05};
level = 0.95; /* this the required alpha value */
sigma = diag(sqrt(1:5)); /* global sigma matrix */
do i = 1 to 4;
m = bm[i,];
plevel = bplevel[i];
geom = (m//(-m))*sigma;
/***************************/
/* Try the null hypothesis */
/***************************/
call seq(prob,geom) eps = 1.e-10;
palpha = (prob[2,]-prob[1,])[5];
/**********************************/
/* Try the alternative hypothesis */
/**********************************/
call seqshift(prob,mean,geom,plevel);
beta = (prob[2,] -prob[1,])[5];
p = prob[3,]-prob[2,]+prob[1,];
/**********************************/
/* Number of patients per group */
/**********************************/
tn = 4*mean##2;
maxn = 5*tn;
/*************************************/
/* compute the average sample number */
/*************************************/
asn = tn *( 5 - (4:0) * p`);
summary = summary // ( palpha || level || beta ||
plevel || tn || maxn ||asn);
end;
print summary[format=10.5];
Note that the variables eps and tscale
have been internally set to their default values.
The following values are returned for the matrix SUMMARY:
These values compare well with the values shown in Table 3 of Pocock (1982). Differences are of the order of
This example shows how to verify the results in Table 1 of Wang and Tsiatis (1987). For a given
start func(delta,k) global(level);
m = ((1:k))##delta;
mm = (-m//m);
/*******************************/
/* meet the significance level */
/* by scaling */
/*******************************/
call seqscale(prob,scale,mm,level);
return(scale);
finish;
/*********************************/
/* alpha levels of 0.05 and 0.01 */
/*********************************/
blevel = {0.95 0.99};
do i = 1 to 2;
level = blevel[i];
free summary;
do delta = 0 to .7 by .1;
free row;
do k=2 to 5;
x = func(delta,k);
row = row || x;
end;
summary = summary //row;
end;
print summary[format=10.5];
end;
The value of SUMMARY for the 0.95 level is as follows.
The value for SUMMARY for the 0.99 level is as follows.
Note that since eps and tscale are not specified, they are internally set to their default values.
This example verifies the results in Table 2 of Pocock (1977). The following program finds
%let nl = 5;
start func(plevel) global(level,scale,mean,palpha,beta,tn,asn);
m = sqrt((1: &nl));
mm = -m //m;
/*******************************/
/* meet the significance level */
/* by scaling */
/*******************************/
call seqscale(prob,scale,mm,level);
palpha = (prob[2,]-prob[1,])[&nl];
mm = mm *scale;
/*******************************/
/* meet the power condition */
/*******************************/
call seqshift(prob,mean,mm,plevel);
return(mean);
finish;
/****************/
/* alpha = 0.95 */
/****************/
level = 9.50000E-01;
bplevel = { 0.5 .25 .1 0.05 0.01};
free summary;
do i = 1 to 5;
summary = summary || func(bplevel[i]);
end;
print summary[format=10.5];
The value returned for SUMMARY are shown in the following
table, and the entries agree with Table 2 of Pocock (1977).
SUMMARY
0.99359 1.31083 1.59229 1.75953 2.07153
This example illustrates how to find the optimal boundary of the
%let nl=5;
start func(delta) global(level,plevel,mean,
scale,alpha,beta,tn,asn);
m = ((1: &nl))##delta;
mm = (-m//m);
/*******************************/
/* meet the significance level */
/*******************************/
call seqscale(prob,scale,mm,level);
alpha = (prob[2,]-prob[1,])[&nl];
mm = mm *scale;
/*******************************/
/* meet the power condition */
/*******************************/
call seqshift(prob,mean,mm,plevel);
beta = (prob[2,]-prob[1,])[&nl];
/*************************************/
/* compute the average sample number */
/*************************************/
p = prob[3,]-prob[2,]+prob[1];
tn = 4*mean##2; /* number per group */
asn = tn *( &nl - p *(%eval(&nl-1):0)`);
return(asn);
finish;
/**********************************************/
/* set up the global variables needed by func */
/**********************************************/
level = 0.95;
plevel = 0.01;
/*****************************************/
/* set up the controlling options of the */
/* optimization routine */
/*****************************************/
opt = {0 2 0 1 6};
tc = repeat(.,1,12);
tc[1] = 100;
tc[7] = 1.e-4;
par = { 1.e-13 . 1.e-10 . . .} || . || epsd;
/*****************************/
/* provide the initial guess */
/* and let nlpdd do the work */
/*****************************/
delta = 0.5;
call nlpdd(rc,rx,"func",delta) opt=opt tc=tc par=par;
The following output displays the results.
Optimization Start
Parameter Estimates
Gradient
Objective
N Parameter Estimate Function
1 X1 -1.500000 -8.09752
Value of Objective Function = 35.232023082
Double Dogleg Optimization
Dual Broyden - Fletcher - Goldfarb - Shanno Update (DBFGS)
Without Parameter Scaling
Gradient Computed by Finite Differences
Number of Parameter Estimates 1
Parameter Estimates 2
Functions (Observations) 2
Optimization Start
Active Constraints 0 Criterion = 35.232
Max Abs Gradient Element 8.098 Radius = 1.000
Function Active Objective
Iter Restart Calls Constraints Function
1 0 3 0 34.8914
2* 0 4 0 34.8774
3* 0 5 0 34.8774
Iter difcrit maxgrad lambda slope
1 0.3406 1.644 49.273 -0.830
2* 0.0140 0.0440 0 -0.0144
3* 0.00001 0.00013 0 -1E-5
Optimization Results
Iterations 3 Function Calls 6
Gradient Calls 5 Active Constraints 0
Criterion 34.877417 Max Grad Element 0.000126832
Slope -0.0000100034 Radius 1
NOTE: FCONV convergence criterion satisfied.
Optimization Results
Parameter Estimates
N Parameter Estimate Gradient
1 X1 0.586554 -0.0001268
Value of Objective Function = 34.877416815
The optimal function value of 34.88 agrees with
the entry in Table 2 of Wang and Tsiatis (1987)
for five groups, ![]() , and
, and ![]() .
Note that the variables eps and tscale
are internally set to their default values.
For more information about the NLPDD subroutine, see the section "NLPDD Call".
For details about the opt, tc, and par
arguments in the NLPDD call, see the section "Options Vector",
the section "Termination Criteria", and
the section "Control Parameters Vector", respectively.
.
Note that the variables eps and tscale
are internally set to their default values.
For more information about the NLPDD subroutine, see the section "NLPDD Call".
For details about the opt, tc, and par
arguments in the NLPDD call, see the section "Options Vector",
the section "Termination Criteria", and
the section "Control Parameters Vector", respectively.
You can replicate other values in Table 2 of Wang and Tsiatis (1987) by changing the values of the variables NL and PLEVEL. You can obtain values from Table 3 by changing the value of the variable LEVEL to 0.99 and specifying NL and PLEVEL accordingly.
This example illustrates how to find the boundaries that minimize ASN given the required significance level and the required power. It replicates some of the results published in Table 3 of Pocock (1982). The IML program computes the domain that
- minimizes the ASN
- yields a given significance level of 0.05
- yields a given power
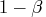 under the alternative hypothesis
under the alternative hypothesis
%let nl=5;
start func(m) global(level,plevel,sigma,epss,
geometry,stgeom,gscale,mean,alpha,beta,tn,asn);
m = abs(m);
mm = ( -m // m)*sigma;
/*******************************/
/* meet the significance level */
/*******************************/
call seqscale(prob,gscale,mm,level) iguess=gscale eps=epss;
stgeom = gscale*m;
geometry= mm*gscale;
alpha = (prob[2,]-prob[1,])[&nl];
/*******************************/
/* meet the power condition */
/*******************************/
call seqshift(prob,mean,geometry,plevel) iguess=mean eps=epss;
beta = (prob[2,]-prob[1,])[&nl];
p = prob[3,] - prob[2,]+prob[1,];
/*************************************/
/* compute the average sample number */
/*************************************/
tn = 4*mean##2; /* number per group */
asn = tn *( &nl - p *(%eval(&nl-1):0)`);
return(asn);
finish;
/**********************************************/
/* set up the global variables needed by func */
/**********************************************/
epss = 1.e-8;
epso = 1.e-5;
level = 9.50000E-01;
plevel = 0.05;
sigma = diag(sqrt(1:5));
/*****************************************/
/* set up the controlling options of the */
/* optimization routine */
/*****************************************/
opt = {0 2 0 1 6};
tc = repeat(.,1,12);
tc[1] = 100;
tc[7] = 1.e-4;
par = { 1.e-13 . 1.e-10 . . .} || . || epso;
/*************************************/
/* provide the constraint matrix */
/* you need monotonically increasing */
/* significance levels */
/*************************************/
con = { . . . . . . . ,
. . . . . . . ,
1 -1 . . . 1 0 ,
. 1 -1 . . 1 0 ,
. . 1 -1 . 1 0 ,
. . . 1 -1 1 0 };
/*****************************/
/* provide the initial guess */
/* and let nlp do the work */
/*****************************/
m = { 1 1 1 1 1 };
call nlpdd(rc,rx,"func",m) opt=opt blc = con tc=tc par=par;
print stgeom;
Note that while eps has been set to eps=Note the following about the optimization process:
- Different levels of precision
are imposed on different modules.
In this example, epss, which is used as the
precision for the sequential tests, is 1E-8.
The absolute and relative function criteria for the
objective function are set to par[7]=1E-5
and tc[7]=1E-4, respectively.
Since finite differences are used to compute the
first and second derivatives, the sequential test
should be more precise than the optimization routine.
Otherwise, the finite difference estimation is worthless.
Optimally, if the precision of the function evaluation
is
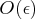 , the first- and second-order
derivatives should be estimated with
perturbations
, the first- and second-order
derivatives should be estimated with
perturbations 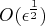 and
and
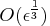 , respectively.
For example, if all three precision levels are set to
1E-5, the optimization process does not work properly.
, respectively.
For example, if all three precision levels are set to
1E-5, the optimization process does not work properly.
- Line search techniques that do not depend on the computation of the derivative are preferable.
- The amount of printed information from the optimization routines is controlled by opt[2] and can be set to any value between 0 and 3, with larger numbers representing more printed output.
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.